Research
-
Disintegration Fingerprinting: A low-cost and easy-to-use tool for identifying substandard and falsified medicines
Ishmam Fatima, Oscar Fajardo, Harshith Sadhu, and William H. Grover, preprint on medRxiv.
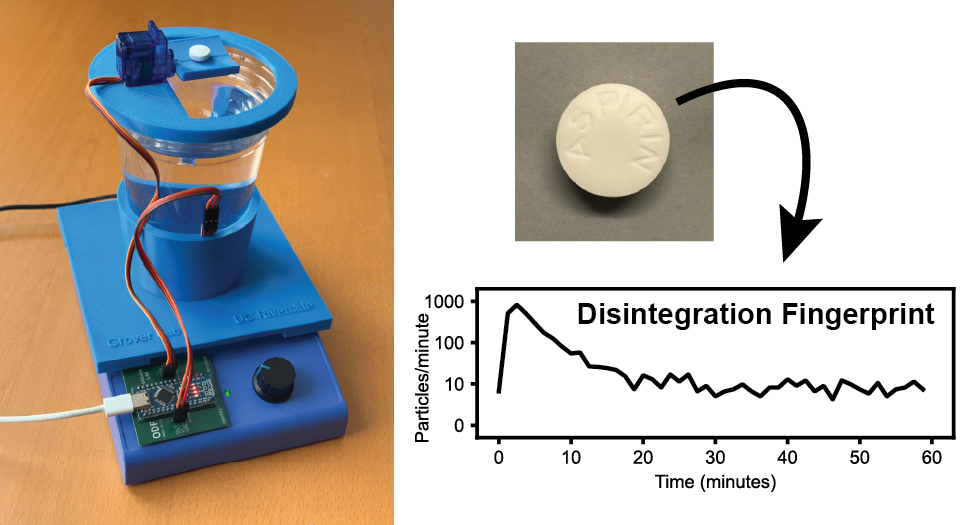
There is an urgent need for low-cost and simple-to-use tools for identifying substandard and falsified medicines. In this work we demonstrate “Disintegration Fingerprinting” (DF), a technique that identifies pills, tablets, caplets, and other solid-dosage drugs based on how the drug disintegrates and dissolves in liquid. The DF hardware consists of a water-filled transparent plastic cup atop a conventional magnetic stirrer. An inexpensive sensor mounted on the outside of the cup shines infrared light into the cup and measures the amount of light that is reflected back to the sensor. When a pill is added to the stirred water, the pill begins to disintegrate into particles that swirl around inside the cup. Whenever one of these particles passes near the infrared sensor, the particle reflects additional light back to the sensor and creates a millisecond-duration peak in a plot of sensor output vs. time. The number of particles in the water changes over time as the particles continue to disintegrate and (in some cases) eventually dissolve away. By plotting the number of particles detected vs. time, we create a Disintegration Fingerprint that can be used to identify the drug product. In a proof-of-concept study, we used DF to analyze 96 pills from 32 different drug products (including antibiotics, opioid and non-opioid analgesics, antidepressants, anti-inflammatories, antiemetics, antihistamines, decongestants, muscle relaxants, expectorants, sleep aids, cold medicines, antacids, hormonal birth control, and dietary supplements, as well as a simulated falsified drug product). We found that DF correctly identified 90% of these pills, and the technique can even distinguish name-brand and generic versions of the same drug. By providing a fast (60-minute), inexpensive ($33 USD), and easy-to-use tool for identifying substandard and falsified medicines, Disintegration Fingerprinting can play an important role in the fight against fake drugs.
-
Wild foundress queen bumble bees make numerous, short foraging trips and exhibit frequent nest failure: Insights from trap-nesting and RFID tracking
Erica Sarro Gustilo, William H. Grover, and S. Hollis Woodard, Ecology and Evolution 15: e71016 (2025).

The overwhelming majority of research on wild bumble bees has focused on the social colony stage. Nest-founding queens in the early season are difficult to study because incipient nests are challenging to find in the wild and the foundress queen flight period is very short relative to the entire nesting period. As a result, natural history information on foundress queens is exceedingly rare. New methodological approaches are needed to adequately study this elusive life stage. We trap-nested wild queen bumble bees in artificial nest boxes in Gothic, Colorado and used a custom-built radio frequency identification (RFID) system to continuously record queen foraging activity (inferred from entering and exiting the nest) for the majority of their spring flight periods. Foundress queens made frequent, short foraging trips, which tended to increase in duration over the course of the flight period. All queens who produced adult workers ceased foraging within approximately 1 week after workers emerged in the nest. We observed frequent nest failure among foundress queens: Fewer than one quarter of queens who laid eggs in nest boxes went on to produce reproductive gynes at the end of the season. We also report nest characteristics and curious phenomena we observed, including conspecific nest invasion and queens remaining outside the nest overnight. We present this trap-nesting and subsequent RFID tracking method as a valuable, albeit resource-intensive, path forward for uncovering new information about the elusive, incipient life stage of wild bumble bees.
-
Controlling biomedical devices using pneumatic logic
Shane Hoang, Mabel Shehada, Konstantinos Karydis, Philip Brisk, and William H. Grover, Annals of Biomedical Engineering 53, 207-216 (2024). PDF
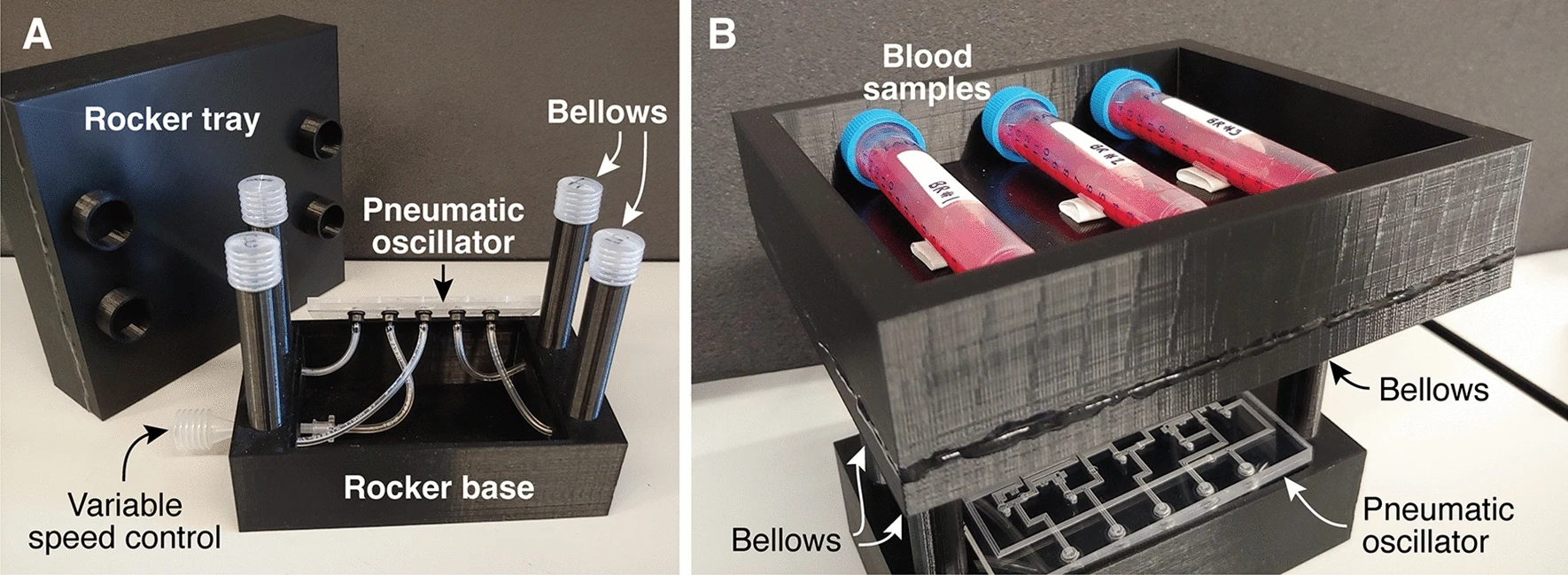
Many biomedical devices are powered and controlled by electrical components. These electronics add to the cost of a device (possibly making the device too expensive for use in resource-limited or point-of-care settings) and can also render the device unsuitable for use in some environments (for example, high-humidity areas such as incubators where condensation could cause electrical short circuits, ovens where electronic components may overheat, or explosive or flammable environments where electric sparks could cause serious accidents). In this work, we show that pneumatic logic can be used to power and control biomedical devices without the need for electricity or electric components. Originally developed for controlling microfluidic “lab-on-a-chip” devices, these circuits use microfluidic valves like transistors in air-powered logic “circuits.” We show that a modification to the basic valve design—adding additional air channels in parallel through the valve—creates a “high-flow” valve that is suitable for controlling a broad range of bioinstruments, not just microfluidics. As a proof-of-concept, we developed a high-flow pneumatic oscillator that uses five high-flow Boolean NOT gates arranged in a loop. Powered by a single constant vacuum source, the oscillator provides five out-of-phase pneumatic outputs that switch between vacuum and atmospheric pressure every 1.3 s. Additionally, a user can adjust the frequency of the oscillator by squeezing a bellows attached to one of the pneumatic outputs. We then used the pneumatic oscillator to power a low-cost 3D-printed laboratory rocker/shaker commonly used to keep blood products, cell cultures, and other heterogeneous samples in suspension. Our air-powered rocker costs around $12 USD to build and performs as well as conventional electronic rockers that cost $1000 USD or more. This is the first of many biomedical devices that can be made cheaper and safer using pneumatic logic.
-
Air-powered logic circuits for error detection in pneumatic systems
Shane Hoang, Mabel Shehada, Zinal Patel, Minh-Huy Tran, Konstantinos Karydis, Philip Brisk, and William H. Grover, Device 2, 100507 (2024).

Pneumatic control systems are common in manufacturing, healthcare, transportation, robotics, and many other fields. Undetected failures in pneumatic systems can have serious consequences. In this work, we present an air-powered error detector that can identify failures in pneumatic systems. This device contains a pneumatic logic circuit of 21 microfluidic valves that calculates the parity bit corresponding to several pneumatic control bits. If a problem such as an air leak or blockage occurs, then the calculated and expected parity bits will not match, and the device outputs an error signal to alert the user or to shut down the system. As a proof of concept, we used the device to detect anomalies in an intermittent pneumatic compression (IPC) medical device. By providing a simple and low-cost way to detect problems without using sensors, the pneumatic error detector can promote safety and reliability across a wide range of pneumatic systems.
-
Multi-objective design automation for microfluidic capture chips
Lisa Chen, William H. Grover, Manu Sridharan, and Philip Brisk, IEEE Transactions on NanoBioscience (2022). PDF
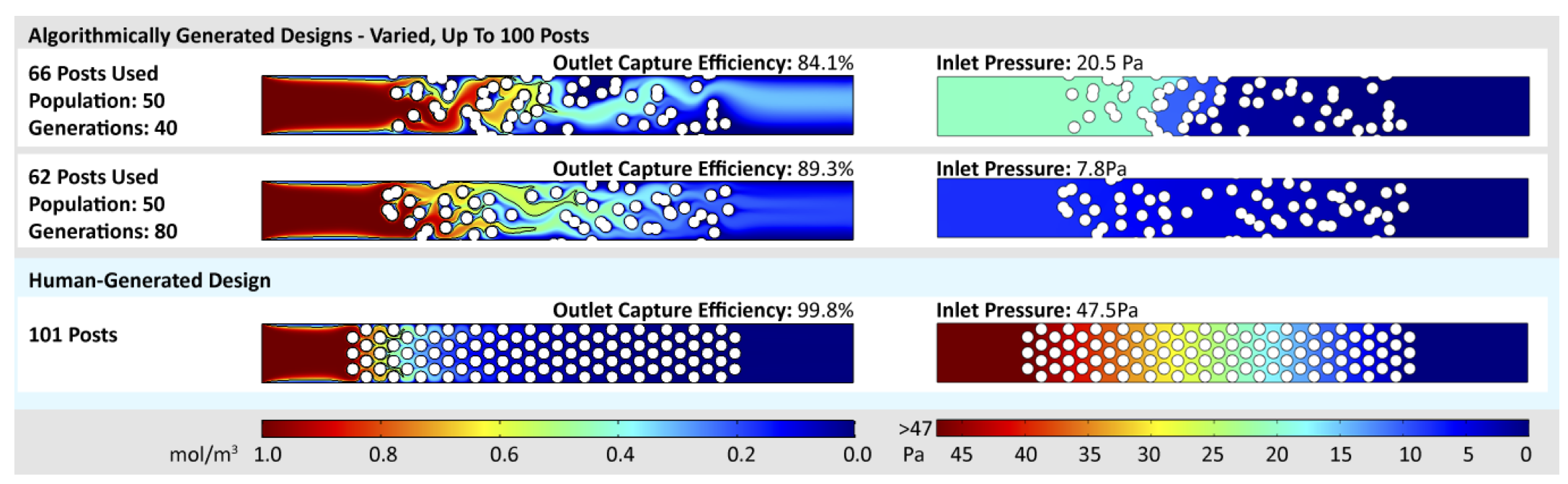
Microfluidic capture chips are useful for preparing or analyzing a wide range of different chemical, biological, and medical samples. A typical microfluidic capture chip contains features that capture certain targets (i.e. molecules, particles, cells) as they flow through the chip. However, creating optimal capture chip designs is difficult because of the inherent relationship between capture efficiency and flow resistance: as more capture features are added to the chip, the capture efficiency increases, but the additional features slow the flow of fluid through the chip. This paper introduces the use of multi-objective optimization to generate capture chip designs that balance the trade-off between maximizing target capture efficiency and minimizing resistance to fluid flow. Design automation for this important class of microfluidic chips has not been attempted previously. Our approach automatically produces a Pareto front of non-dominated chip designs in a reasonable amount of time, and most of these designs have comparable capture efficiency to hand-designed chips with far lower flow resistance. By choosing from the chip designs on the Pareto front, a user can obtain high capture efficiency without exceeding the flow resistance constraints of their application.
-
CandyCodes: simple universally unique edible identifiers for confirming the authenticity of pharmaceuticals
William H. Grover, Scientific Reports 12: 7452 (2022)
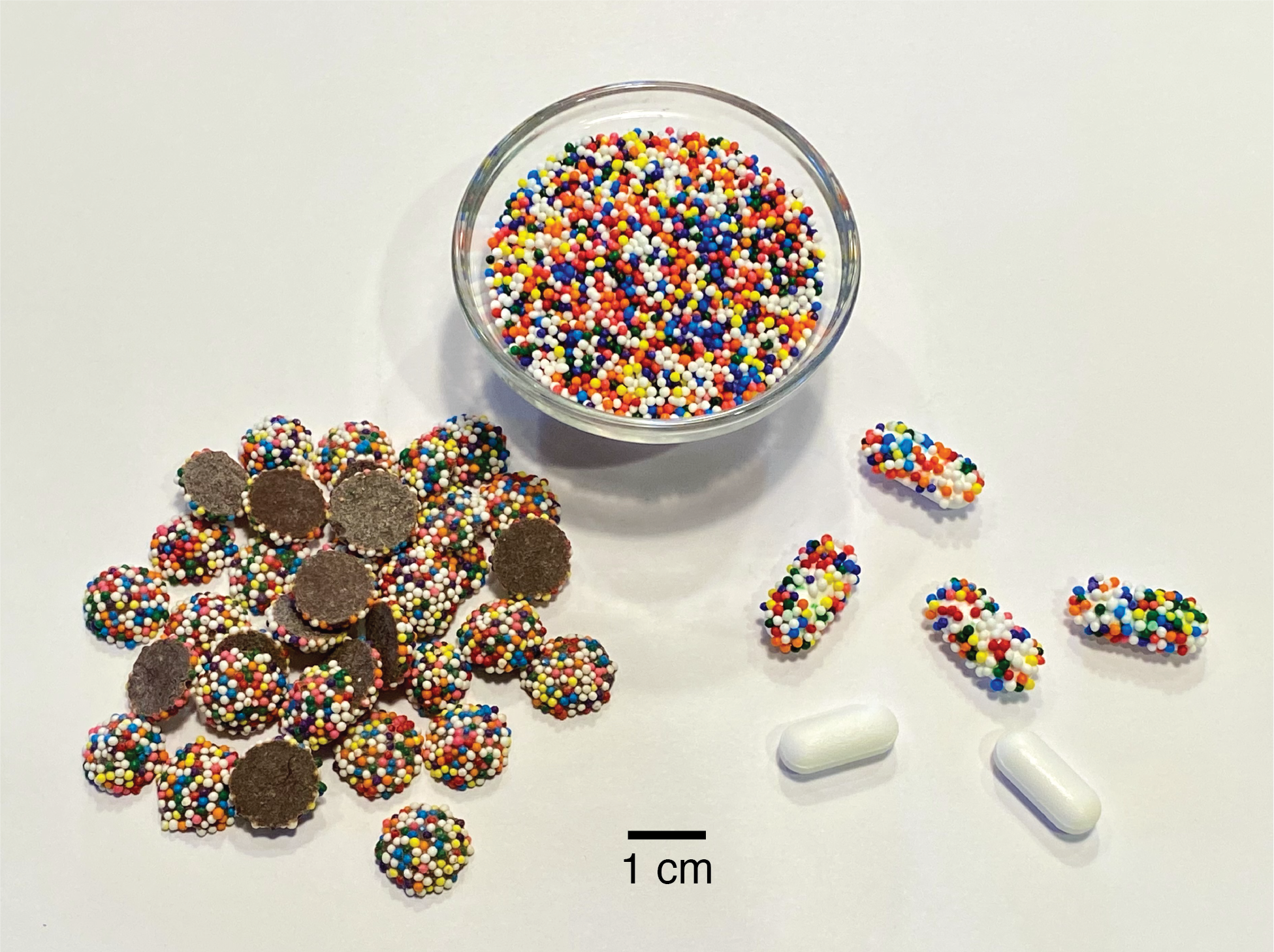
Counterfeit or substandard medicines adversely affect the health of millions of people and cost an estimated $200 billion USD annually. Their burden is greatest in developing countries, where the World Health Organization estimates that one in ten medical products are fake. In this work, I describe a simple addition to the existing drug manufacturing process that imparts an edible universally unique physical identifier to each pill, tablet, capsule, caplet, etc. This technique uses nonpareils (also called sprinkles and “hundreds and thousands”), tiny inexpensive multicolor candy spheres that are normally added to other candies or desserts as decorations. If nonpareils are applied at random to a pill immediately after manufacture, the specific pattern they form is unlikely to ever be repeated by random chance; this means that the pattern (or “CandyCode”) can be used to uniquely identify the pill and distinguish it from all other pills. By taking a photograph of each CandyCoded pill after manufacture and recording the location and color of each nonpareil, a manufacturer can construct a database containing the CandyCodes of all known-authentic pills they produce. A consumer can then simply use a cellphone to photograph a pill and transfer its image to the manufacturer’s server, which determines whether the pill’s CandyCode matches a known-good CandyCode in their database (meaning that the pill is authentic) or does not have a match in the database (in which case the consumer is warned that the pill may be counterfeit and should not be consumed). To demonstrate the feasibility of using random particles as universal identifiers, I performed a series of experiments using both real CandyCodes (on commercially produced chocolate candies) and simulated CandyCodes (generated by software). I also developed a simple method for converting a CandyCode photo to a set of strings for convenient storage and retrieval in a database. Even after subjecting CandyCodes to rough handling to simulate shipping conditions, the CandyCodes were still easily verifiable using a cellphone camera. A manufacturer could produce at least 10^17 CandyCoded pills—41 million for each person on Earth—and still be able to uniquely identify each CandyCode. By providing universally-unique IDs that are easy to manufacture but hard to counterfeit, require no alteration of the existing drug formulation and minimal alteration of the manufacturing process, and need only a cameraphone for verification, CandyCodes could play an important role in the fight against fraud in pharmaceuticals and many other products.
-
A pneumatic random-access memory for controlling soft robots
Shane Hoang, Konstantinos Karydis, Philip Brisk, and William H. Grover, PLOS ONE 16 (7): e0254524 (2021).
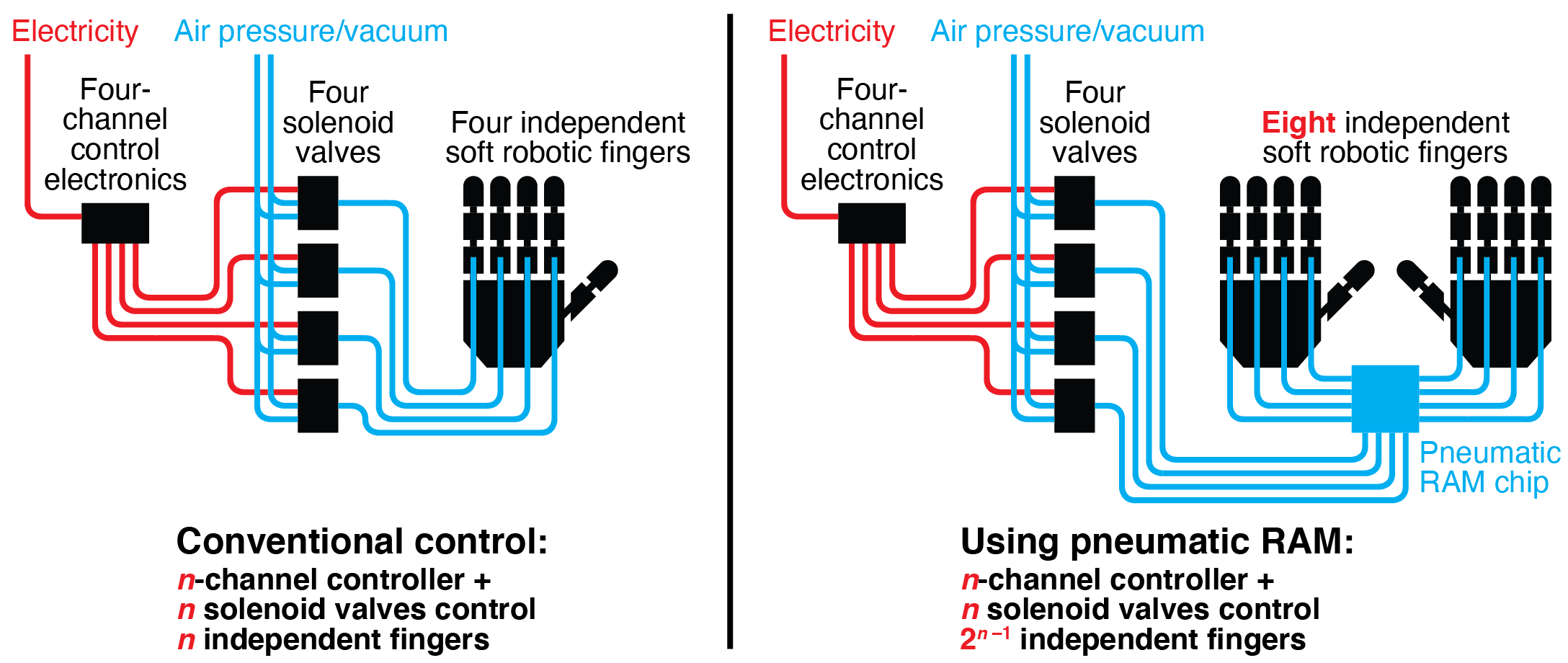
Pneumatically-actuated soft robots have advantages over traditional rigid robots in many applications. In particular, their flexible bodies and gentle air-powered movements make them more suitable for use around humans and other objects that could be injured or damaged by traditional robots. However, existing systems for controlling soft robots currently require dedicated electromechanical hardware (usually solenoid valves) to maintain the actuation state (expanded or contracted) of each independent actuator. When combined with power, computation, and sensing components, this control hardware adds considerable cost, size, and power demands to the robot, thereby limiting the feasibility of soft robots in many important application areas. In this work, we introduce a pneumatic memory that uses air (not electricity) to set and maintain the states of large numbers of soft robotic actuators without dedicated electromechanical hardware. These pneumatic logic circuits use normally-closed microfluidic valves as transistor-like elements; this enables our circuits to support more complex computational functions than those built from normally-open valves. We demonstrate an eight-bit nonvolatile random-access pneumatic memory (RAM) that can maintain the states of multiple actuators, control both individual actuators and multiple actuators simultaneously using a pneumatic version of time division multiplexing (TDM), and set actuators to any intermediate position using a pneumatic version of analog-to-digital conversion. We perform proof-of-concept experimental testing of our pneumatic RAM by using it to control soft robotic hands playing individual notes, chords, and songs on a piano keyboard. By dramatically reducing the amount of hardware required to control multiple independent actuators in pneumatic soft robots, our pneumatic RAM can accelerate the spread of soft robotic technologies to a wide range of important application areas.
-
Measuring dissolution profiles of single controlled-release drug pellets
Heran C. Bhakta, Jessica M. Lin, and William H. Grover, Scientific Reports 10, 19734 (2020) .
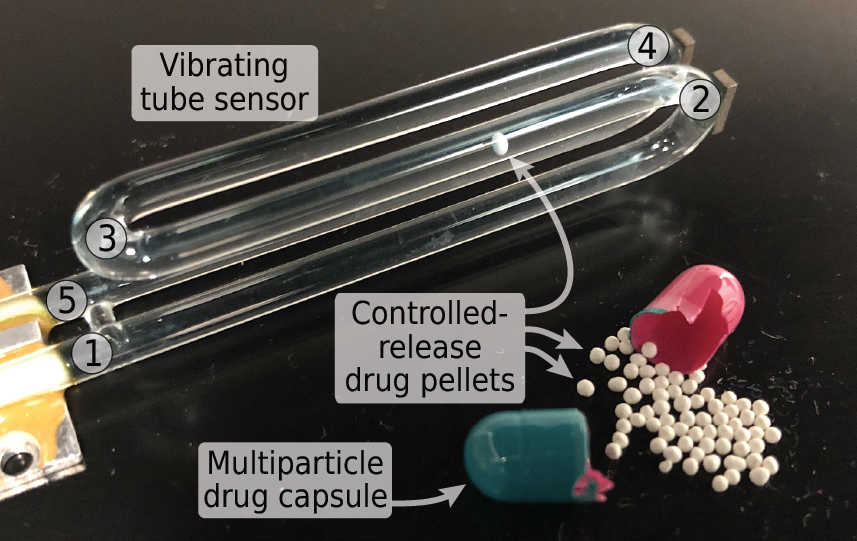
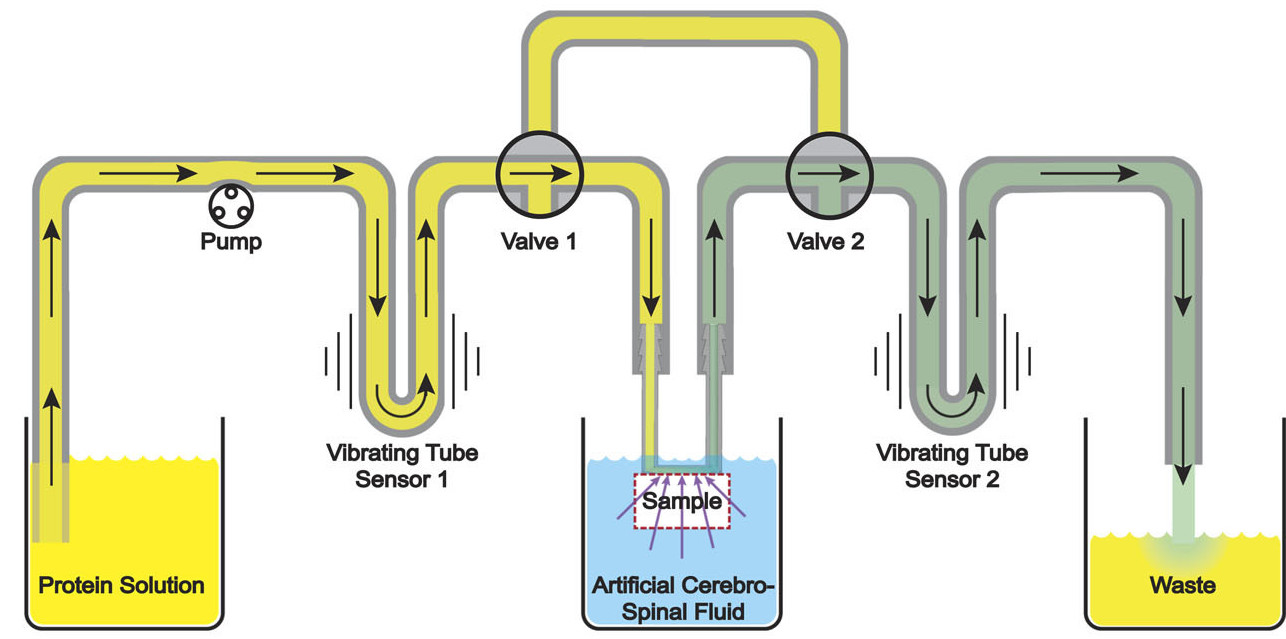
Many solid-dose oral drug products are engineered to release their active ingredients into the body at a certain rate. Techniques for measuring the dissolution or degradation of a drug product in vitro play a crucial role in predicting how a drug product will perform in vivo. However, existing techniques are often labor-intensive, time-consuming, irreproducible, require specialized analytical equipment, and provide only “snapshots” of drug dissolution every few minutes. These limitations make it difficult for pharmaceutical companies to obtain full dissolution profiles for drug products in a variety of different conditions, as recommended by the US Food and Drug Administration. Additionally, for drug dosage forms containing multiple controlled-release pellets, particles, beads, granules, etc. in a single capsule or tablet, measurements of the dissolution of the entire multi-particle capsule or tablet are incapable of detecting pellet-to-pellet variations in controlled release behavior. In this work, we demonstrate a simple and fully-automated technique for obtaining dissolution profiles from single controlled-release pellets. We accomplished this by inverting the drug dissolution problem: instead of measuring the increase in the concentration of drug compounds in the solution during dissolution (as is commonly done), we monitor the decrease in the buoyant mass of the solid controlled-release pellet as it dissolves. We weigh single controlled-release pellets in fluid using a vibrating tube sensor, a piece of glass tubing bent into a tuning-fork shape and filled with any desired fluid. An electronic circuit keeps the glass tube vibrating at its resonance frequency, which is inversely proportional to the mass of the tube and its contents. When a pellet flows through the tube, the resonance frequency briefly changes by an amount that is inversely proportional to the buoyant mass of the pellet. By passing the pellet back-and-forth through the vibrating tube sensor, we can monitor its mass as it degrades or dissolves, with high temporal resolution (measurements every few seconds) and mass resolution (700 nanogram resolution). As a proof-of-concept, we used this technique to measure the single-pellet dissolution profiles of several commercial controlled-release proton pump inhibitors in simulated stomach and intestinal contents, as well as comparing name-brand and generic formulations of the same drug. In each case, vibrating tube sensor data revealed significantly different dissolution profiles for the different drugs, and in some cases our method also revealed differences between different pellets from the same drug product. By measuring any controlled-release pellets, particles, beads, or granules in any physiologically-relevant environment in a fully-automated fashion, this method can augment and potentially replace current dissolution tests and support product development and quality assurance in the pharmaceutical industry.
-
ChemStor: Using Formal Methods To Guarantee Safe Storage and Disposal of Chemicals
Jason Ott, Daniel Tan, Tyson Loveless, William H. Grover, and Philip Brisk, Journal of Chemical Information and Modeling 60 (7), 3416-3422 (2020).
While safe chemical storage and disposal are simple in principle—users should read safety specifications and place chemicals in appropriate cabinets or collection points—high-profile incidents involving improper storage and disposal of chemicals continue to occur. This paper introduces ChemStor, an open-source, automated computational system that can guarantee (mathematically verify a system is correct with respect to its specification), with regard to prescribed constraints, safe storage and disposal of chemicals used in academic, industrial, and domestic settings. ChemStor borrows concepts from formal methods—a branch of computer science capable of mathematically proving a specification or software is correct—to safely store or dispose of chemicals. If two or more chemicals can be combined in the same cabinet without forming possibly dangerous combinations of chemicals (while observing cabinet/shelf space constraints), then ChemStor determines that the storage configuration is safe. Likewise, if chemicals can be added to an existing disposal container without forming possibly dangerous combinations of chemicals (or exceeding the volume of the container), then ChemStor determines that the disposal configuration is safe. ChemStor accomplishes this by first building a chemical interaction graph, a graph that describes which chemicals may interact with each other based on their reactivity groups as determined by the United States Environmental Protection Agency. Next, ChemStor computes the chromatic number of the graph, the smallest number of colors used to color the graph such that no two vertices (chemicals) that share an edge (an interaction) share the same color. ChemStor then assigns all the chemicals of each color to a storage or disposal container after confirming that there is enough space in the container. These steps are encoded into a series of satisfiability modulo theory equations, and ChemStor uses an industry-standard tool to try to find a valid solution to these equations. The result is either a solution which dictates exactly where to store or dispose of each chemical, or an indication that no safe storage or disposal configuration could be found. To demonstrate the feasibility of ChemStor, we used the tool to analyze ten real-world chemical storage and disposal incidents that led to injuries or destruction of property. In each case, ChemStor quickly and successfully identified a proper chemical disposal or storage configuration that would have prevented the incident. In the future, ChemStor may be integrated with electronic laboratory notebooks, voice assistants, and other emerging technology to protect users of chemicals in labs, workplaces, and homes.
-
Differential densimetry: A method for determining ultra-low fluid flux and tissue permeability
Christopher S. Hale, Heran C. Bhakta, Carrie R. Jonak, Jennifer M. Yonan, Devin K. Binder, William H. Grover, and Victor J.G. Rodgers, AIP Advances 9, 095063 (2019).
Osmotic transport devices (OTDs) are forward osmosis membrane devices that we recently developed to remove accumulated fluid from swollen tissue, in-vivo, under severe conditions. As such, the relative volume of the fluid required to be removed and the volumetric flowrate may be two orders of magnitude less than the operating volume and tangential flowrate of the device. This makes it challenging to measure the rate of fluid flow from the swollen tissue. Here, we introduce a differential densimetry method for determining ultra-low volumetric flux through tissue samples. This technique uses two vibrating tube density sensors, one placed upstream of the membrane in contact with the tissue sample, and one placed downstream. Any flow of biological fluid withdrawn through the tissue will combine with the OTD operating fluid resulting in an observed density shift in the second density sensor. By measuring the difference in density between the upstream and downstream fluids, one can calculate the amount of fluid flowing across the tissue with a relatively high level of sensitivity. This method is also relatively insensitive to drift from temperature fluctuations and capable of continuously monitoring tissue permeability in real time. As a proof of concept, we used this technique to measure fluid flow across ex-vivo rat spinal tissue for an appropriately scaled OTD. The repeatability error had a maximum of only 12%. This implies that this method can provide highly acceptable flux measurements with reasonable reproducibility in real-time applications of fluid removal in-vivo.
-
Finding the optimal design of a passive microfluidic mixer
Junchao Wang, Naiyin Zhang, Jin Chen, Victor G.J. Rodgers, Philip Brisk, and William H. Grover, Lab on a Chip 19, 3618–3627 (2019). PDF
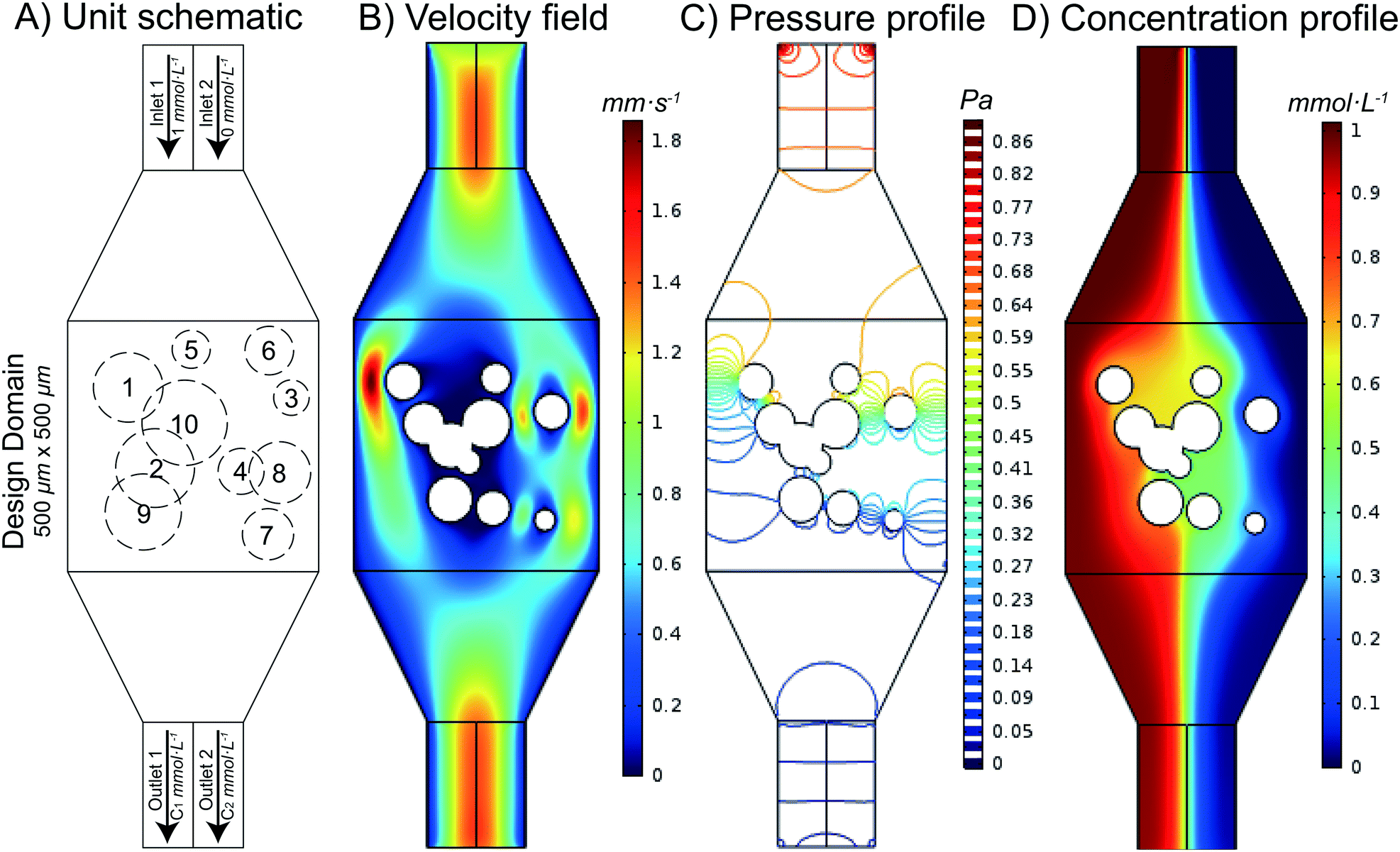
The ability to thoroughly mix two fluids is a fundamental need in microfluidics. While a variety of different microfluidic mixers have been designed by researchers, it remains unknown which (if any) of these mixers are optimal (that is, which designs provide the most thorough mixing with the smallest possible fluidic resistance across the mixer). In this work, we automatically designed and rationally optimized a microfluidic mixer. We accomplished this by first generating a library of thousands of different randomly designed mixers, then using the non-dominated sorting genetic algorithm II (NSGA-II) to optimize the random chips in order to achieve Pareto efficiency. Pareto efficiency is a state of allocation of resources (e.g. driving force) from which it is impossible to reallocate so as to make any one individual criterion better off (e.g. pressure drop) without making at least one individual criterion (e.g. mixing performance) worse off. After 200 generations of evolution, Pareto efficiency was achieved and the Pareto-optimal front was found. We examined designs at the Pareto-optimal front and found several design criteria that enhance the mixing performance of a mixer while minimizing its fluidic resistance; these observations provide new criteria on how to design optimal microfluidic mixers. Additionally, we compared the designs from NSGA-II with some popular microfluidic mixer designs from the literature and found that designs from NSGA-II have lower fluidic resistance with similar mixing performance. As a proof of concept, we fabricated three mixer designs from 200 generations of evolution and one conventional popular mixer design and tested the performance of these four mixers. Using this approach, an optimal design of a passive microfluidic mixer is found and the criteria of designing a passive microfluidic mixer are established.
-
Using printer ink color to control the behavior of paper microfluidics
Joshua Potter, Philip Brisk, and William H. Grover, Lab on a Chip 19, 2000-2008 (2019).
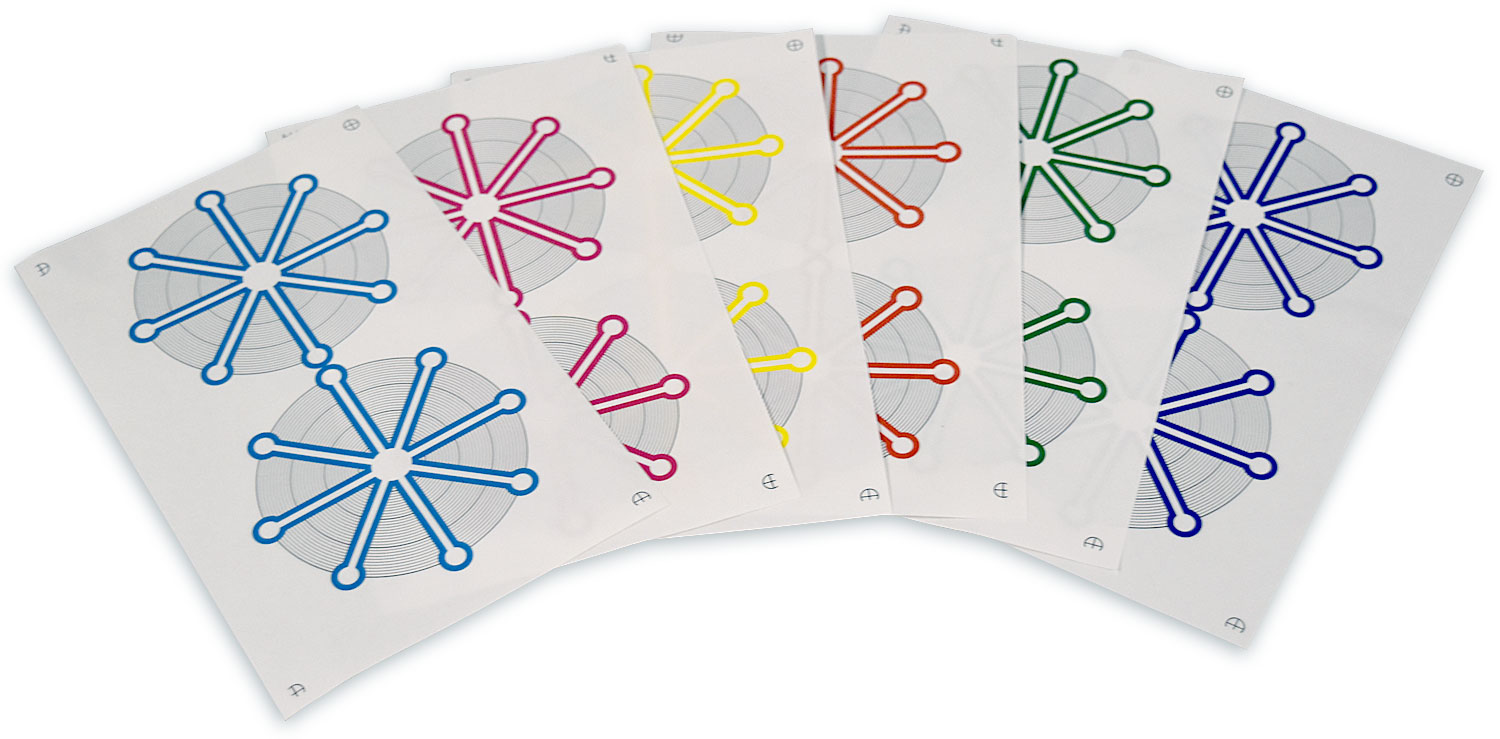
Paper microfluidic devices (including lateral flow assays) offer an excellent combination of utility and low cost. Many paper microfluidic devices are fabricated using the Xerox ColorQube line of commercial wax-based color printers; the wax ink serves as a hydrophobic barrier to fluid flow. These printers are capable of depositing four different colors of ink, cyan (C), magenta (M), yellow (Y), and black (K), plus 11 combinations of these colors (CM, CY, CK, MY, MK, YK, CMY, CMK, CYK, MYK, and CMYK), although most researchers use only black ink to print paper microfluidic devices. Recently, as part of a project to develop a computer-aided design framework for use with paper microfluidics devices, we unexpectedly observed that different colors of wax ink behave differently in paper microfluidics. We found that among the single colors of ink, black ink actually had the most barrier failures, and magenta ink had the fewest barrier failures. In addition, some combinations of colors performed even better than magenta: the combinations CY, MK, YK, CMY, CYK and MYK had no barrier failures in our study. We also found that the printer delivers significantly different amounts of ink to the paper for the different color combinations, and in general, the color combinations that formed the strongest barriers to fluid flow were the ones that had the most ink delivered to the paper. This suggests that by simply weighing paper samples printed with all 15 combinations of colors, one can easily find the color combinations most likely to form a strong barrier for a given printer. Finally, to show that deliberate choices of ink colors can actually be used to create new functions in paper microfluidics, we designed and tested a new color-based “antifuse” structure that protects paper microfluidic devices from a typical operator error (addition of too much fluid to the device). Our results provide a set of color choice guidelines that designers can use to control the behavior of their paper microfluidics.
-
Chronoprints: Identifying samples by visualizing how they change over space and time
Brittney A. McKenzie, Jessica Robles-Najar, Eric Duong, Philip Brisk, and William H. Grover, ACS Central Science 5 (4), 589–598 (2019).
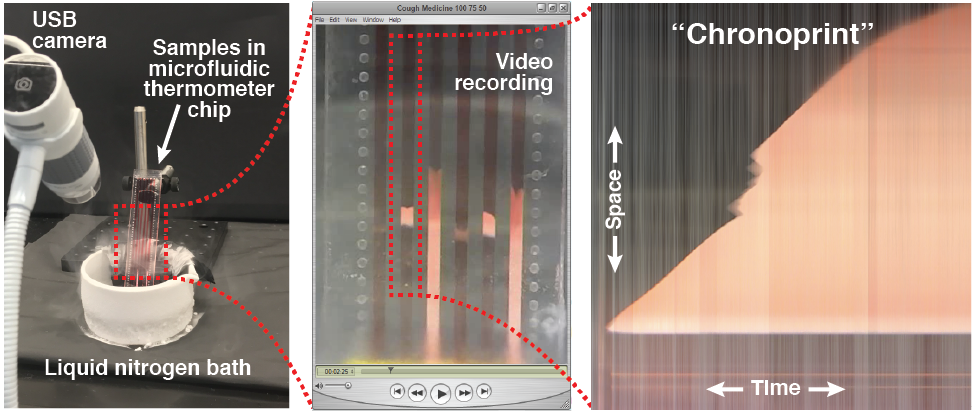
The modern tools of chemistry excel at identifying a sample, but the cost, size, complexity, and power consumption of these instruments often preclude their use in resource-limited settings. In this work, we demonstrate a simple and low-cost method for identifying a sample based on visualizing how the sample changes over space and time in response to a perturbation. Different types of perturbations could be used, and in this proof-of-concept we use a dynamic temperature gradient that rapidly cools different parts of the sample at different rates. We accomplish this by first loading several samples into long parallel channels on a “microfluidic thermometer chip.” We then immerse one end of the chip in liquid nitrogen to create a dynamic temperature gradient along the channels, and we use an inexpensive USB microscope to record a video of how the samples respond to the changing temperature gradient. The video is then converted into several bitmap images (one per sample) that capture each sample’s response to the perturbation in both space (the y-axis; the distance along the dynamic temperature gradient) and time (the x-axis); we call these images “chronological fingerprints” or “chronoprints” of each sample. If two samples’ chronoprints are similar, this suggests that the samples are the same chemical substance or mixture, but if two samples’ chronoprints are significantly different, this proves that the samples are chemically different. Since chronoprints are just bitmap images, they can be compared using a variety of techniques from computer science, and in this work we use three different image comparison algorithms to quantify chronoprint similarity. As a demonstration of the versatility of chronoprints, we use them in three different applications: distinguishing authentic olive oil from adulterated oil (an example of the over $10 billion global problem of food fraud), identifying adulterated or counterfeit medication (which represents around 10% of all medication in low- and middle-income countries), and distinguishing the occasionally-confused pharmaceutical ingredients glycerol and diethylene glycol (whose accidental or intentional substitution has led to hundreds of deaths). The simplicity and versatility of chronoprints should make them valuable analytical tools in a variety of different fields.
-
Oligo-Snoop: A Non-Invasive Side Channel Attack Against DNA Synthesis Machines
Sina Faezi, Sujit Rokka Chhetri, Arnav Vaibhav Malawade, John Charles Chaput, William Grover, Philip Brisk, and Mohammad Abdullah Al Faruque, 2019 Network and Distributed Systems Security (NDSS) Symposium, San Diego, CA.
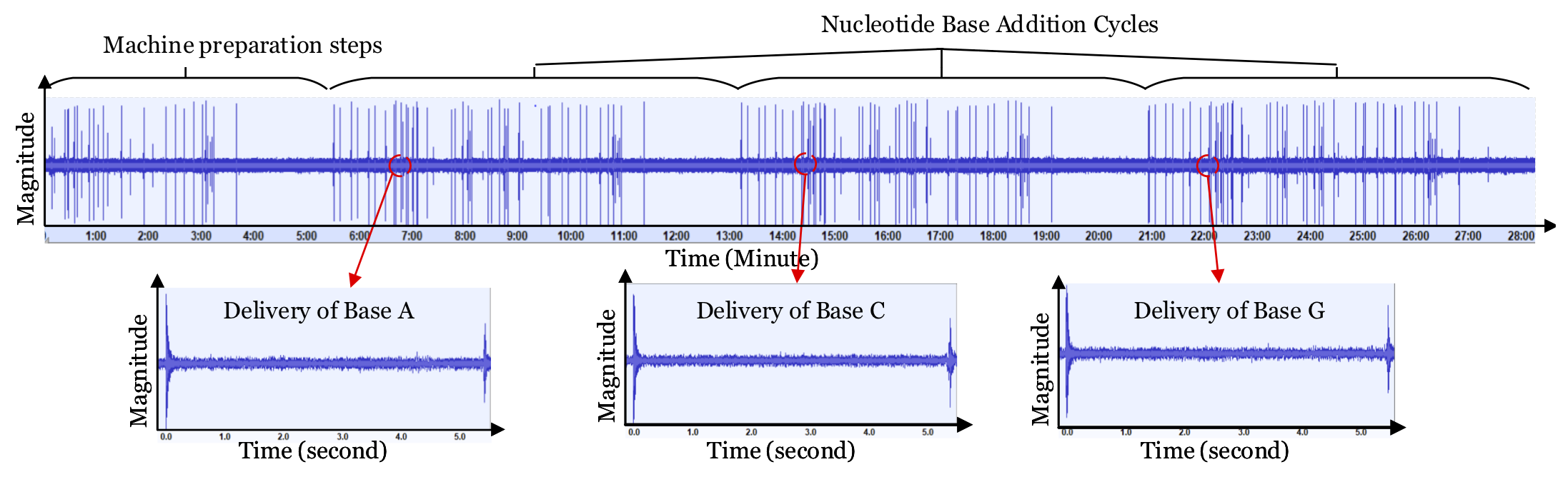
Synthetic biology is developing into a promising science and engineering field. One of the enabling technologies for this field is the DNA synthesizer. It allows researchers to custom-build sequences of oligonucleotides (short DNA strands) using the nucleobases: Adenine (A), Guanine (G), Cytosine (C), and Thymine (T). Incorporating these sequences into organisms can result in improved disease resistance and lifespan for plants, animals, and humans. Hence, many laboratories spend large amounts of capital researching and developing unique sequences of oligonucleotides. However, these DNA synthesizers are fully automated systems with cyber-domain processes and physical domain components. Hence, they may be prone to security breaches like any other computing system. In our work, we present a novel acoustic side-channel attack methodology which can be used on DNA synthesizers to breach their confidentiality and steal valuable oligonucleotide sequences. Our proposed attack methodology achieves an average accuracy of 88.07% in predicting each base and is able to reconstruct short sequences with 100% accuracy by making less than 21 guesses out of 415 possibilities. We evaluate our attack against the effects of the microphone’s distance from the DNA synthesizer and show that our attack methodology can achieve over 80% accuracy when the microphone is placed as far as 0.7 meters from the DNA synthesizer despite the presence of common room noise. In addition, we reconstruct DNA sequences to show how effectively an attacker with biomedical-domain knowledge would be able to derive the intended functionality of the sequence using the proposed attack methodology. To the best of our knowledge, this is the first methodology that highlights the possibility of such an attack on systems used to synthesize DNA molecules.
-
Musical instruments as sensors
Heran C. Bhakta, Vamsi K. Choday, and William H. Grover. ACS Omega 3 (9), 11026-11032 (2018).
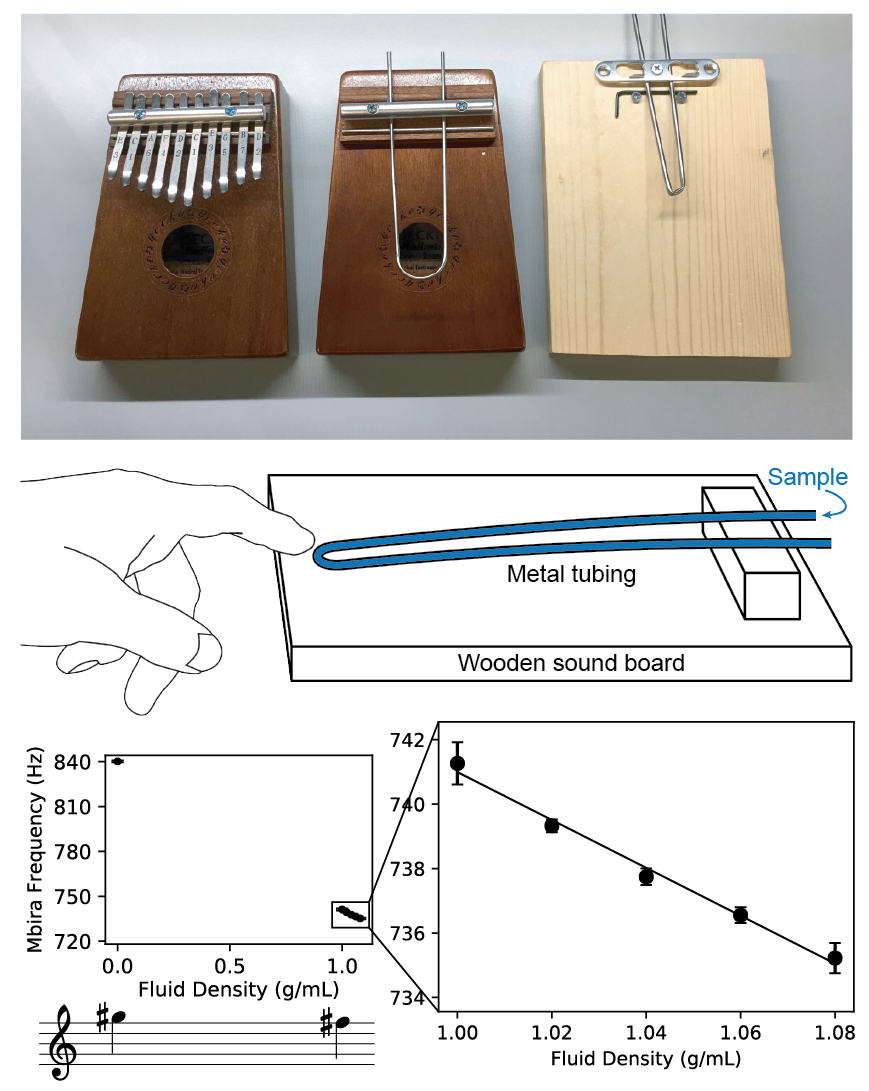
The frequencies of notes made by a musical instrument are determined by the physical properties of the instrument. Consequently, by measuring the frequency of a note, one can infer information about the instrument’s physical properties. In this work, we show that by modifying a musical instrument to contain a sample and analyzing the instrument’s pitch, we can make precision measurements of the physical properties of the sample. We used the mbira, a 3000-year-old African musical instrument that consists of metal tines attached to a wooden board; these tines are plucked to play musical notes. By replacing the mbira’s tines with bent steel tubing, filling the tubing with a sample, using a smartphone to record the sound while plucking the tubing, and measuring the frequency of the sound using a free software tool on our website, we can measure the density of the sample with a resolution of about 0.012 g/mL. Unlike existing tools for measuring density, the mbira sensor can be made and used by virtually anyone in the world. To demonstrate the mbira sensor’s capabilities, we used it to successfully distinguish diethylene glycol and glycerol, two similar chemicals that are sometimes mistaken for each other in pharmaceutical manufacturing (leading to hundreds of deaths). We also show that consumers could use mbira sensors to detect counterfeit and adulterated medications (which represent around 10% of all medications in low- and middle-income countries). We expect that many other musical instruments can function as sensors and find important and lifesaving applications.
-
A microfluidic thermometer: Precise temperature measurements in microliter- and nanoliter-scale volumes
Brittney A. McKenzie and William H. Grover. PLOS ONE 12 (12): e0189430 (2017).
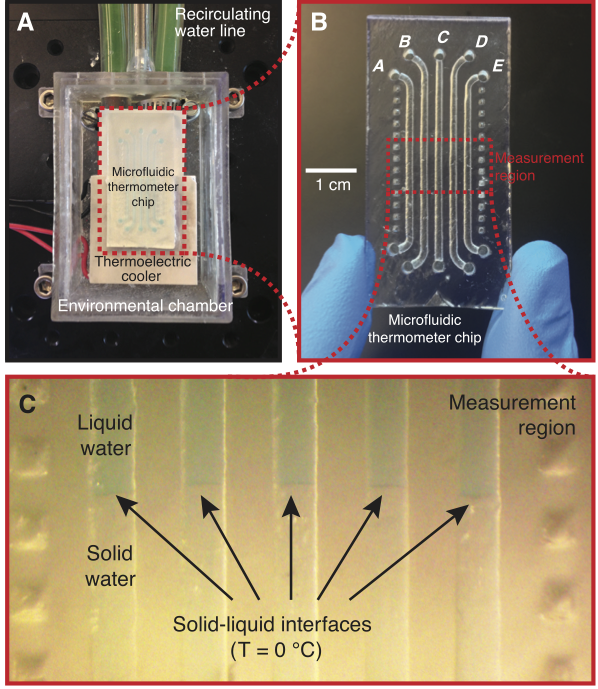
Measuring the temperature of a sample is a fundamental need in many biological and chemical processes. When the volume of the sample is on the microliter or nanoliter scale (e.g., cells, microorganisms, precious samples, or samples in microfluidic devices), accurate measurement of the sample temperature becomes challenging. In this work, we demonstrate a technique for accurately determining the temperature of microliter volumes using a simple 3D-printed microfluidic chip. We accomplish this by first filling “microfluidic thermometer” channels on the chip with substances with precisely known freezing/melting points. We then use a thermoelectric cooler to create a stable and linear temperature gradient along these channels within a measurement region on the chip. A custom software tool (available as online Supporting Information) is then used to find the locations of solid-liquid interfaces in the thermometer channels; these locations have known temperatures equal to the freezing/melting points of the substances in the channels. The software then uses the locations of these interfaces to calculate the temperature at any desired point within the measurement region. Using this approach, the temperature of any microliter-scale on-chip sample can be measured with an uncertainty of about a quarter of a degree Celsius. As a proof-of-concept, we use this technique to measure the unknown freezing point of a 50 microliter volume of solution and demonstrate its feasibility on a 400 nanoliter sample. Additionally, this technique can be used to measure the temperature of any on-chip sample, not just near-zero-Celsius freezing points. We demonstrate this by using an oil that solidifies near room temperature (coconut oil) in a microfluidic thermometer to measure on-chip temperatures well above zero Celsius. By providing a low-cost and simple way to accurately measure temperatures in small volumes, this technique should find applications in both research and educational laboratories.
-
Instantaneous simulation of fluids and particles in complex microfluidic devices
Junchao Wang, Victor G. J. Rodgers, Philip Brisk, and William H. Grover. PLOS ONE 12 (12): e0189429 (2017).
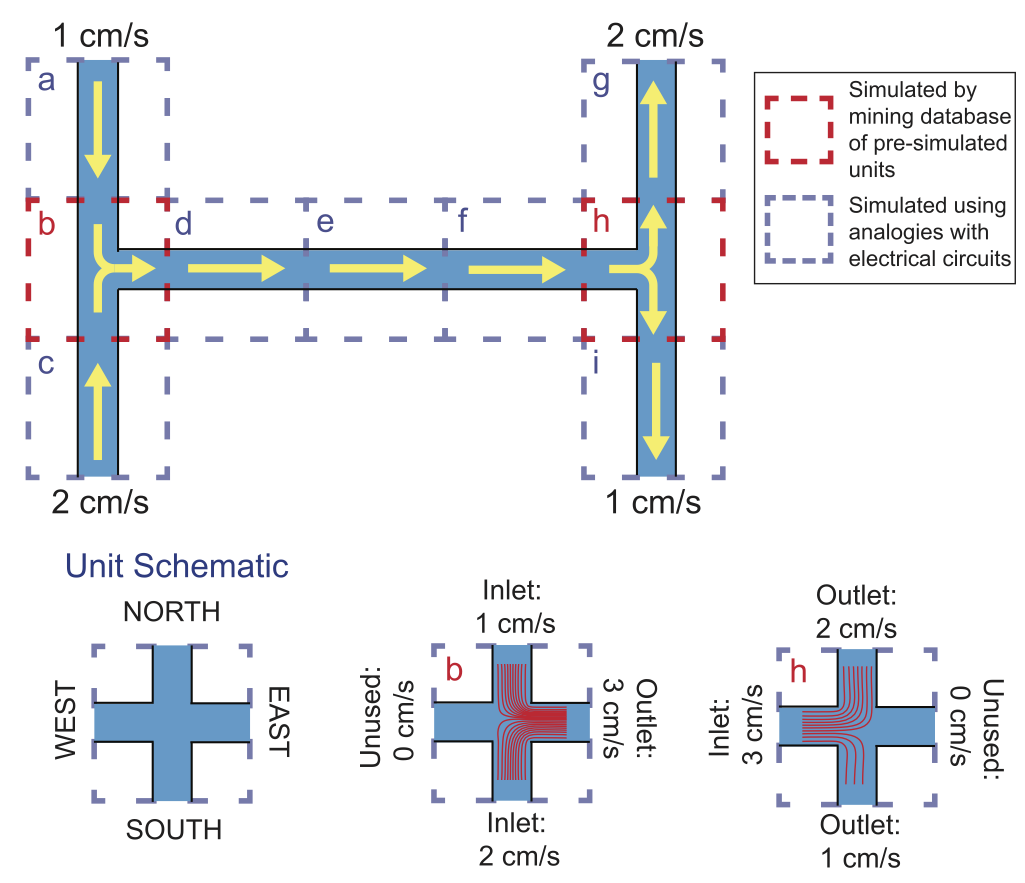
Microfluidics researchers are increasingly using computer simulation in many different aspects of their research. However, these simulations are often computationally intensive: simulating the behavior of a simple microfluidic chip can take hours to complete on typical computing hardware, and even powerful workstations can lack the computational capabilities needed to simulate more complex chips. This slows the development of new microfluidic chips for new applications. To address this issue, we present a microfluidic simulation method that can simulate the behavior of fluids and particles in some typical microfluidic chips instantaneously (in around one second). Our method decomposes the chip into its primary components: channels and intersections. The behavior of fluid in each channel is determined by leveraging analogies with electronic circuits, and the behavior of fluid and particles in each intersection is determined by querying a database containing nearly 100,000 pre-simulated channel intersections. While constructing this database takes a nontrivial amount of computation time, once built, this database can be queried to determine the behavior of fluids and particles in a given intersection in a fraction of a second. Using this approach, the behavior of a microfluidic chip can be simulated in just one second on a standard laptop computer, without any noticeable degradation in the accuracy of the simulation. While our current technique has some constraints on the designs of the chips it can simulate (namely, T- or cross-shaped intersections, 90 degree channel turns, a fixed channel width, fluid flow rates between 0 and 2 cm/s, and particles with diameters between 1 and 20 microns), we provide several strategies for increasing the range of possible chip designs that can be simulated using our technique. As a proof of concept, we show that our simulation method can instantaneously simulate the paths followed by particles in both simple and complex microfluidic chips, with results that are essentially indistinguishable from simulations that took hours or even days to complete using conventional approaches.
-
Sorting cells by their density
Nazila Norouzi, Heran C. Bhakta, and William H. Grover. PLOS ONE 12 (7): e0180520 (2017).
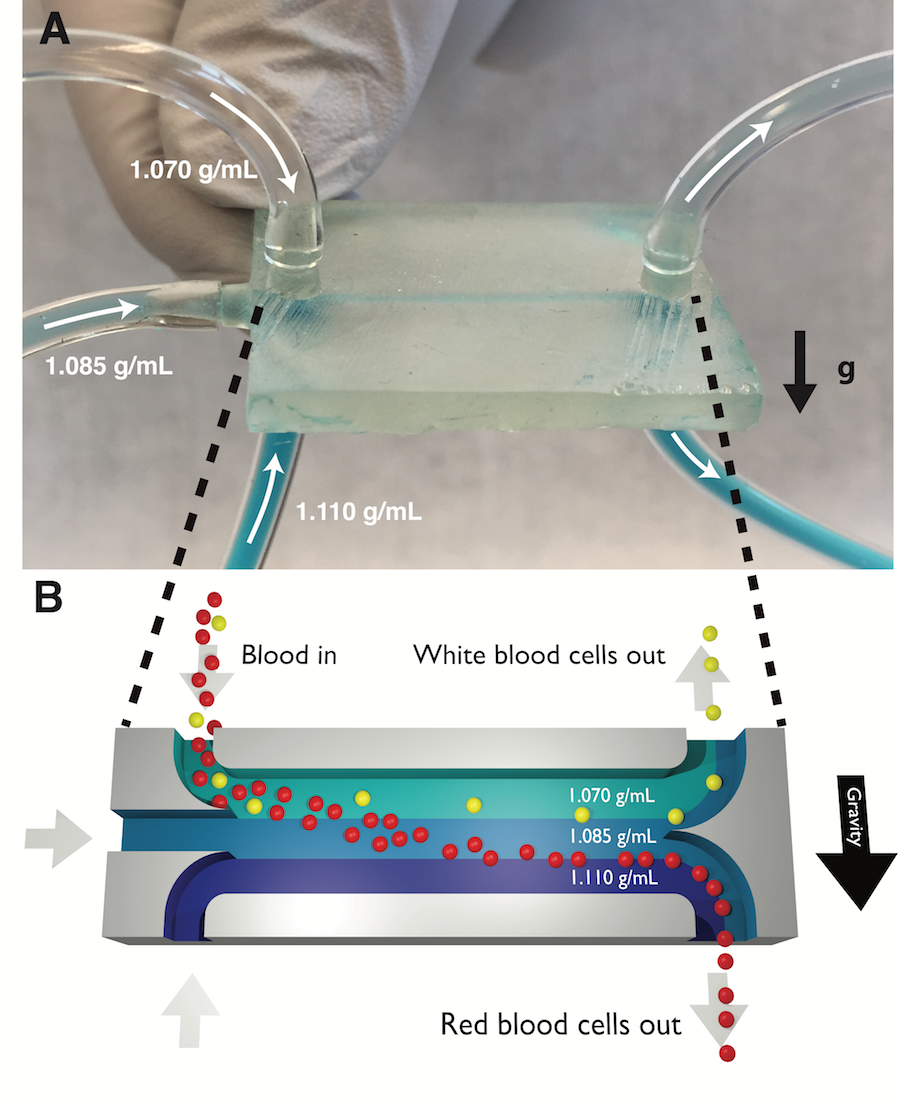
Sorting cells by their type is an important capability in biological research and medical diagnostics. However, most cell sorting techniques rely on labels or tags, which may have limited availability and specificity. Sorting different cell types by their different physical properties is an attractive alternative to labels because all cells intrinsically have these physical properties. But some physical properties, like cell size, vary significantly from cell to cell within a cell type; this makes it difficult to identify and sort cells based on their sizes alone. In this work we continuously sort different cells types by their density, a physical property with much lower cell-to-cell variation within a cell type (and therefore greater potential to discriminate different cell types) than other physical properties. We accomplish this using a 3D-printed microfluidic chip containing a horizontal flowing micron-scale density gradient. As cells flow through the chip, Earth’s gravity makes each cell move vertically to the point where the cell’s density matches the surrounding fluid’s density. When the horizontal channel then splits, cells with different densities are routed to different outlets. As a proof of concept, we use our density sorter chip to sort polymer microbeads by their material (polyethylene and polystyrene) and blood cells by their type (white blood cells and red blood cells). The chip enriches the fraction of white blood cells in a blood sample from 0.1% (in whole blood) to nearly 98% (in the output of the chip), a 1000x enrichment. Any researcher with access to a 3D printer can easily replicate our density sorter chip and use it in their own research using the design files provided as online Supporting Information. Additionally, researchers can simulate the performance of a density sorter chip in their own applications using the Python-based simulation software that accompanies this work. The simplicity, resolution, and throughput of this technique make it suitable for isolating even rare cell types in complex biological samples, in a wide variety of different research and clinical applications.
-
MOPSA: A microfluidics-optimized particle simulation algorithm
Junchao Wang, Victor G.J. Rodgers, Philip Brisk, and William H. Grover. Biomicrofluidics 11 (3): 034121 (2017). PDF
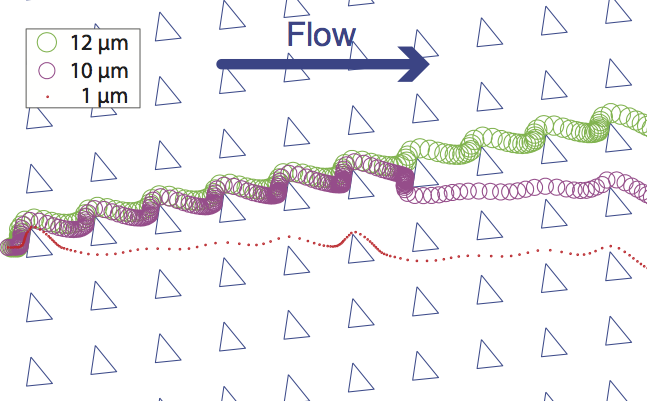
Computer simulation plays a growing role in the design of microfluidic chips. However, the particle tracers in some existing commercial computational fluid dynamics software are not well suited for accurately simulating the trajectories of particles such as cells, microbeads, and droplets in microfluidic systems. To address this issue, we present a microfluidics-optimized particle simulation algorithm (MOPSA) that simulates the trajectories of cells, droplets, and other particles in microfluidic chips with more lifelike results than particle tracers in existing commercial software. When calculating the velocity of a particle, MOPSA treats the particle as a two-dimensional rigid circular object instead of a single point. MOPSA also checks for unrealistic interactions between particles and channel walls and applies an empirical correcting function to eliminate these errors. To validate the performance of MOPSA, we used it to simulate a variety of important features of microfluidic devices like channel intersections and deterministic lateral displacement (DLD) particle sorter chips. MOPSA successfully predicted that different particle sizes will have different trajectories in six published DLD experiments from three research groups; these DLD chips were used to sort a variety of different cells, particles, and droplets. While some of these particles are not actually rigid or spherical, MOPSA’s approximation of these particles as rigid spheres nonetheless resulted in lifelike simulations of the behaviors of these particles (at least for the particle sizes and types shown here). In contrast, existing commercial software failed to replicate these experiments. Finally, to demonstrate that MOPSA can be extended to simulate other properties of particles, we added support for simulating particle density to MOPSA and then used MOPSA to simulate the operation of a microfluidic chip capable of sorting cells by their density. By enabling researchers to accurately simulate the behavior of some types of particles in microfluidic chips before fabricating the chips, MOPSA should accelerate the development of new microfluidic devices for important applications.
-
Design automation for paper microfluidics with passive flow substrates
Joshua Potter, William H. Grover, and Philip Brisk. 27th Annual ACM Great Lakes Symposium on VLSI (GLSVLSI), 215–220 (2017). PDF
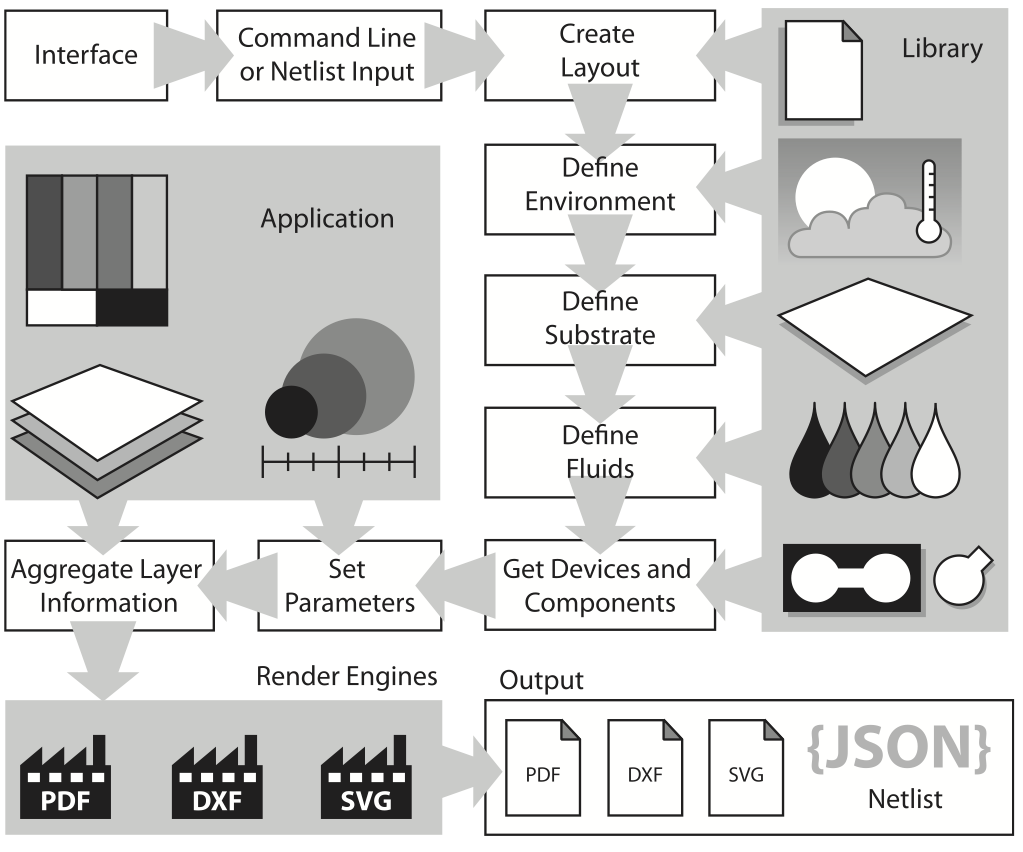
This paper introduces a novel software framework to support automated development of paper-based microfluidic devices. Compared to existing lab-on-a-chip technologies, paper-based microfluidics differs in terms of substrate technologies and point-of-care usage across a wide variety environmental conditions. This paper addresses the contexts in which the software can address these challenges and presents several initial case studies that demonstrate the capabilities of the framework to produce workable and usable paper microfluidic devices.
-
Measuring the mass, volume, and density of microgram-sized objects in fluid
Shirin Mesbah Oskui, Heran C. Bhakta, Graciel Diamante, Huinan Liu, Daniel Schlenk, and William H. Grover. PLOS ONE 12 (4): e0174068 (2017). PDF
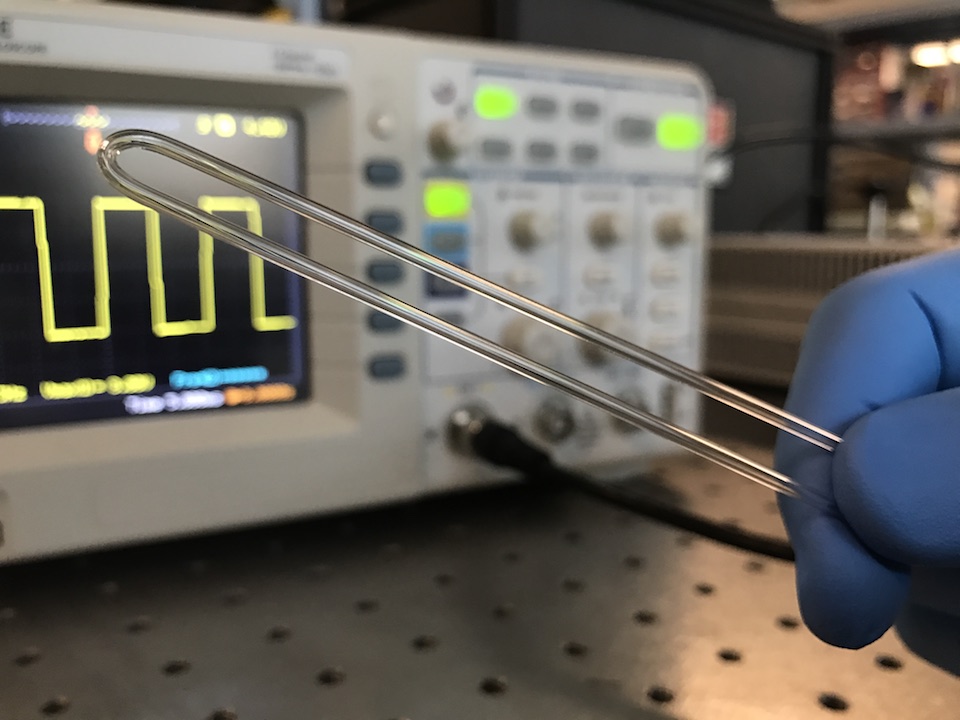
Measurements of an object’s fundamental physical properties like mass, volume, and density can offer valuable insights into the composition and state of the object. However, many important biological samples reside in a liquid environment where it is difficult to accurately measure their physical properties. We show that by using a simple piece of glass tubing and some inexpensive off-the-shelf electronics, we can create a sensor that can measure the mass, volume, and density of microgram-sized biological samples in their native liquid environment. As a proof-of-concept, we use this sensor to measure mass changes in zebrafish embryos reacting to toxicant exposure, density changes in seeds undergoing rehydration and germination, and degradation rates of biomaterials used in medical implants. Since all objects have these physical properties, this sensor has immediate applications in a wide variety of different fields including developmental biology, toxicology, materials science, plant science, and many others.
-
The Case for Semi-Automated Design of Microfluidic Very Large Scale Integration (mVLSI) Chips
Jeffrey McDaniel, William H. Grover, and Philip Brisk. Design, Automation and Test in Europe (DATE) 1793–1798 (2017). PDF
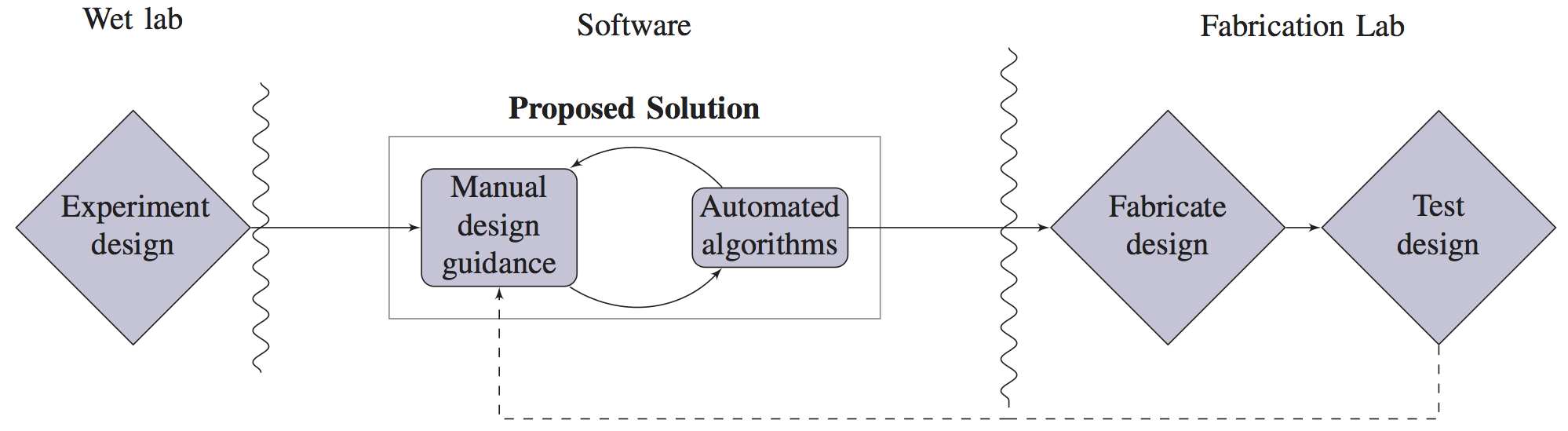
In recent years, significant interest has emerged in the problem of fully automating the design of microfluidic very large scale integration (mVLSI) chips, a popular class of Lab-on-a-Chip (LoC) devices that can automatically execute a wide variety of biological assays. To date, this work has been carried out with little to no input from LoC designers. We conducted interviews with approximately 100 LoC designers, biologists, and chemists from academia and industry; uniformly, they expressed frustration with existing design solutions, primarily commercially available software such as AutoCAD and Solidworks; however, they expressed limited interest and considerable skepticism about the potential for “push-button” end-to-end automation. In response, we have developed a semi-automated mVLSI drawing tool that is designed specifically to address the pain points elucidated by our interviewees. We have used this tool to rapidly reproduce several previously published LoC architectures and generate fabrication ready specifications.
-
Random design of microfluidics
Junchao Wang, Philip Brisk, and William H. Grover. Lab on a Chip 16, 4212-4219 (2016). PDF
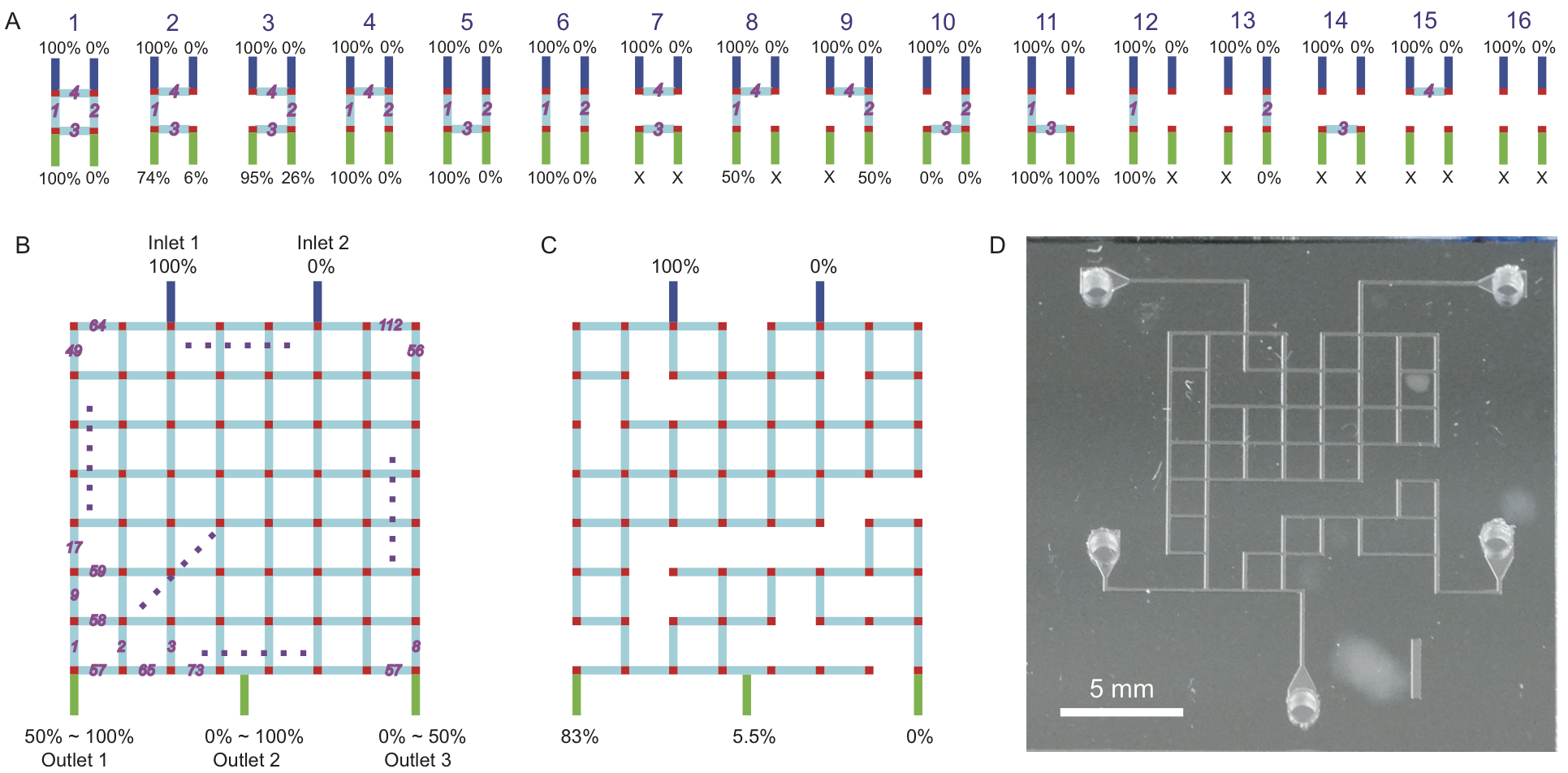
In this work we created functional microfluidic chips without actually designing them. We accomplished this by first generating a library of thousands of different random microfluidic chip designs, then simulating the behavior of each design on a computer using automated finite element analysis. The simulation results were then saved to a database which a user can query via a public website (http://random.groverlab.org) to find chip designs suitable for a specific task. To demonstrate this functionality, we used our library to select chip designs that generate any three desired concentrations of a solute. We also fabricated and tested 16 chips from the library, confirmed that they function as predicted, and used these chips to perform a cell growth rate assay. This is one of many different applications for randomly-designed microfluidics; in principle, any microfluidic chip that can be simulated could be designed automatically using our method. Using this approach, individuals with no training in microfluidics can obtain custom chip designs for their own unique needs in just a few seconds.
-
MECs: "Building Blocks" for Creating Biological and Chemical Instruments
Douglas A. Hill, Lindsey E. Anderson, Casey J. Hill, Afshin Mostaghim, Victor G. J. Rodgers, and William H. Grover. PLOS ONE 11 (7): e0158706. PDF
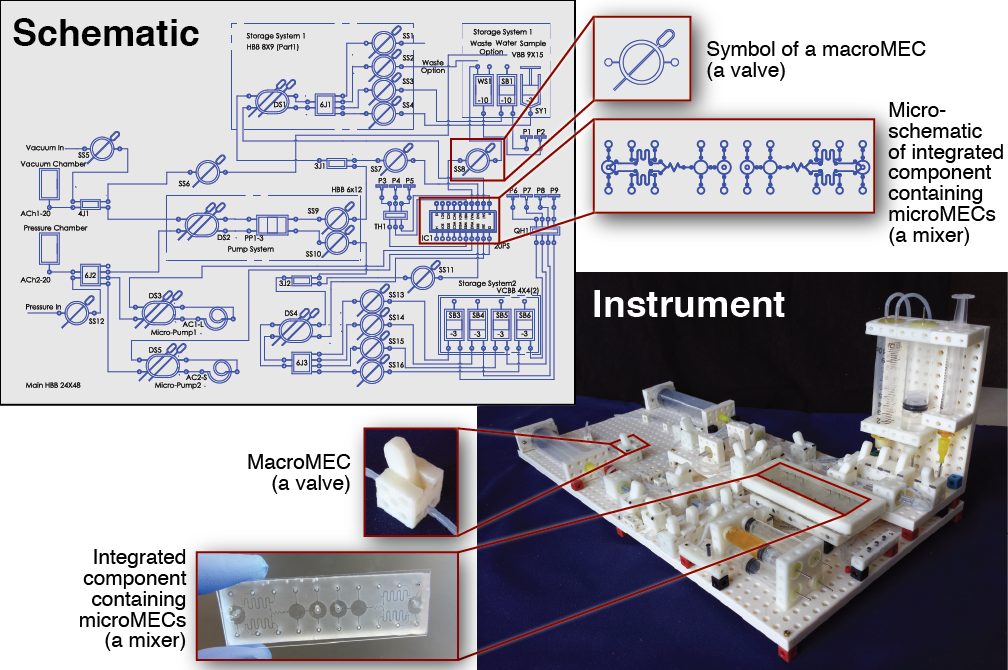
The development of new biological and chemical instruments for research and diagnostic applications is often slowed by the cost, specialization, and custom nature of these instruments. New instruments are built from components that are drawn from a host of different disciplines and not designed to integrate together, and once built, an instrument typically performs a limited number of tasks and cannot be easily adapted for new applications. Consequently, the process of inventing new instruments is very inefficient, especially for researchers or clinicians in resource-limited settings. To improve this situation, we propose that a family of standardized multidisciplinary components is needed, a set of “building blocks” that perform a wide array of different tasks and are designed to integrate together. Using these components, scientists, engineers, and clinicians would be able to build custom instruments for their own unique needs quickly and easily. In this work we present the foundation of this set of components, a system we call Multifluidic Evolutionary Components (MECs). “Multifluidic” conveys the wide range of fluid volumes MECs operate upon (from nanoliters to milliliters and beyond); “multi” also reflects the multiple disciplines supported by the system (not only fluidics but also electronics, optics, and mechanics). “Evolutionary” refers to the design principles that enable the library of MEC parts to easily grow and adapt to new applications. Each MEC “building block” performs a fundamental function that is commonly found in biological or chemical instruments, functions like valving, pumping, mixing, controlling, and sensing. Each MEC also has a unique symbol linked to a physical definition, which enables instruments to be designed rapidly and efficiently using schematics. As a proof-of-concept, we use MECs to build a variety of instruments, including a fluidic routing and mixing system capable of manipulating fluid volumes over five orders of magnitude, an acid-base titration instrument suitable for use in schools, and a bioreactor suitable for maintaining and analyzing cell cultures in research and diagnostic applications. These are the first of many instruments that can be built by researchers, clinicians, and students using the MEC system.
-
Orientation-based control of microfluidics
Nazila Norouzi, Heran C. Bhakta, and William H. Grover. PLOS ONE 11 (3): e0149259. PDF
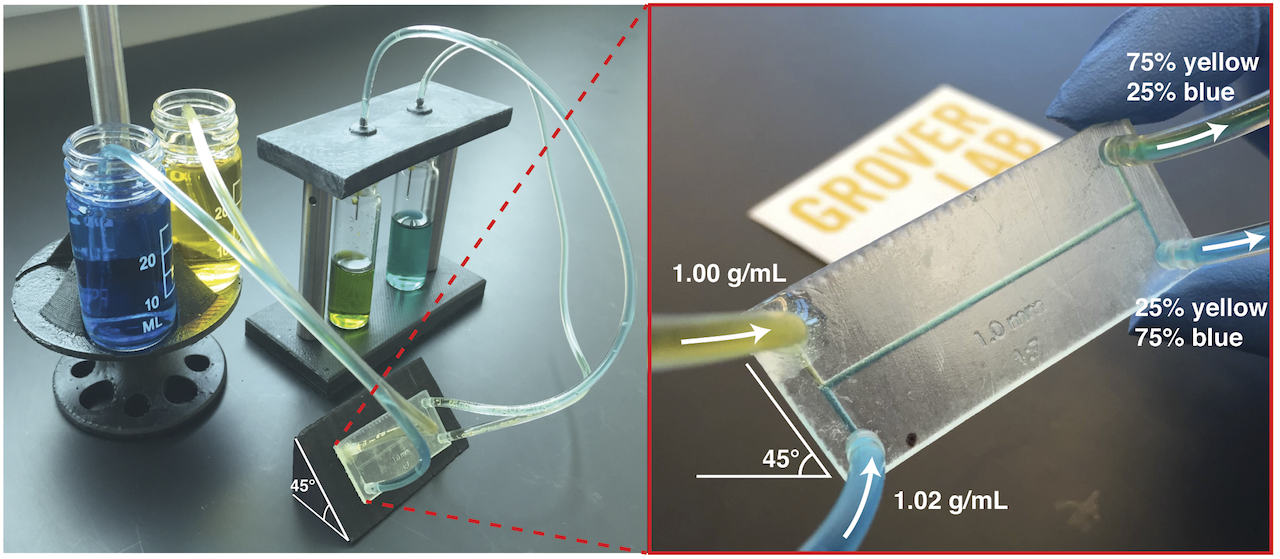
Most microfluidic chips utilize off-chip hardware (syringe pumps, computer-controlled solenoid valves, pressure regulators, etc.) to control fluid flow on-chip. This expensive, bulky, and power-consuming hardware severely limits the utility of microfluidic instruments in resource-limited or point-of-care contexts, where the cost, size, and power consumption of the instrument must be limited. In this work, we present a technique for on-chip fluid control that requires no off-chip hardware. We accomplish this by using inert compounds to change the density of one fluid in the chip. If one fluid is made 2% more dense than a second fluid, when the fluids flow together under laminar flow the interface between the fluids quickly reorients to be orthogonal to Earth’s gravitational force. If the channel containing the fluids then splits into two channels, the amount of each fluid flowing into each channel is precisely determined by the angle of the channels relative to gravity. Thus, any fluid can be routed in any direction and mixed in any desired ratio on-chip simply by holding the chip at a certain angle. This approach allows for sophisticated control of on-chip fluids with no off-chip control hardware, significantly reducing the cost of microfluidic instruments in point-of-care or resource-limited settings.
-
Cyber-physical integration in programmable microfluidic biochips
Tsung-Yi Ho, William H. Grover, Shiyan Hu, and Krishnendu Chakrabarty. 33rd IEEE International Conference on Computer Design (ICCD), 224-227 (2015). PDF
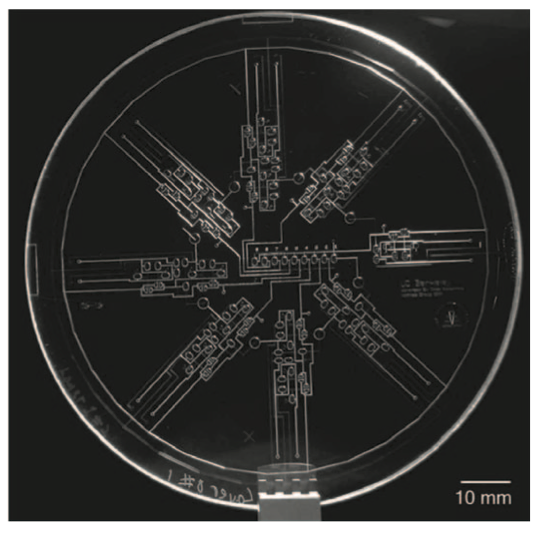
Microfluidic biochip technology integrates miniaturized components into a chip that can perform traditional biochemical laboratory procedures. Commercial impact is highlighted by the recent acquisition of Advanced Liquid Logic by Illumina Inc., a leader in DNA sequencing and biomolecular analysis. Due to the inherent variability involved in many biochemical processes, uncertainties manifest themselves in many ways in microfluidics. Cyber-physical integration of on-chip sensors permits feedback-driven monitoring in-real time to detect and correct errors, along with other benefits such as adaptive control and dynamic re-synthesis. This paper overviews flow-based and digital (droplet-based) microfluidic biochips, and discusses the state-of-the-art in microfluidic device fabrication, the interplay between sensor feedback and adaptive control software, and practical experiences relating to biochip cyber-physical integration. It demonstrates the connections between the many fundamental principles of chip design and engineering, and the needs of the biochip community.
-
Flow-layer physical design for microchips based on monolithic membrane valves
Jeffrey McDaniel, Brian Crites, Philip Brisk, and William H. Grover. IEEE Design and Test 32 (6), 51-59 (2015). PDF
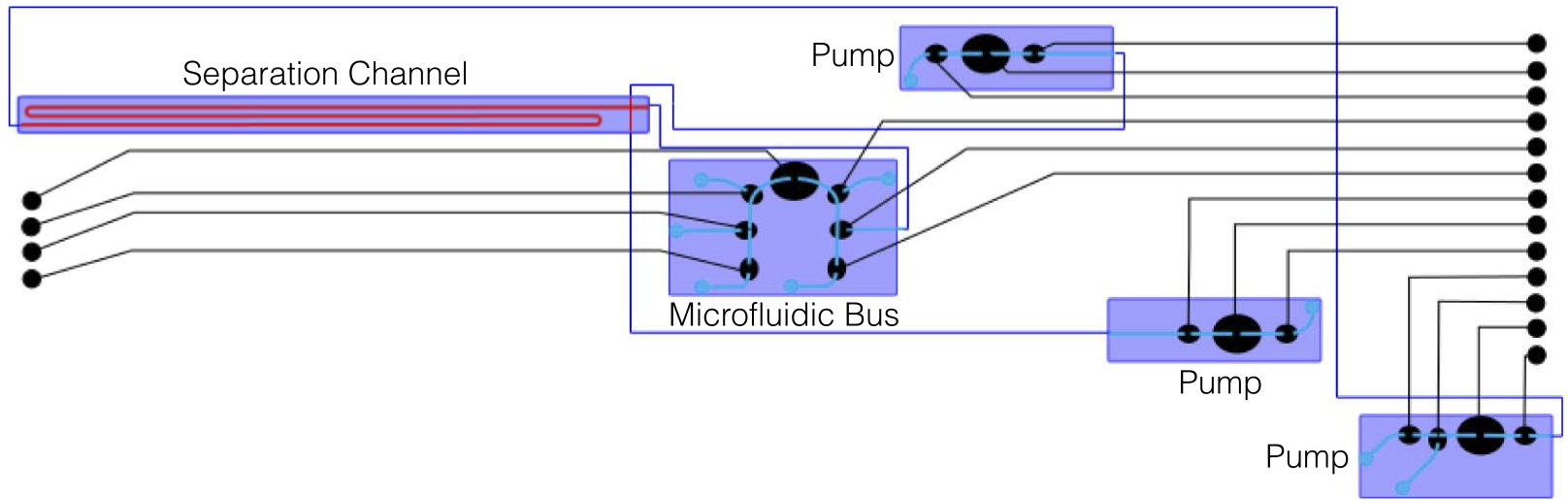
This article introduces a software toolchain for physical design and layout for the flow layer of microfluidic LoCs based on integrated microvalve technology. A case study shows that it can automatically produce layouts for the Mars Organic Analyzer LoC to detect biomolecules in soil on Mars.
-
Assessing and Reducing the Toxicity of 3D-Printed Parts
Shirin Mesbah Oskui, Graciel Diamante, Chunyang Liao, Wei Shi, Jay Gan, Daniel Schlenk, and William H. Grover, Environmental Science and Technology Letters 3 (1): 1–6. PDF
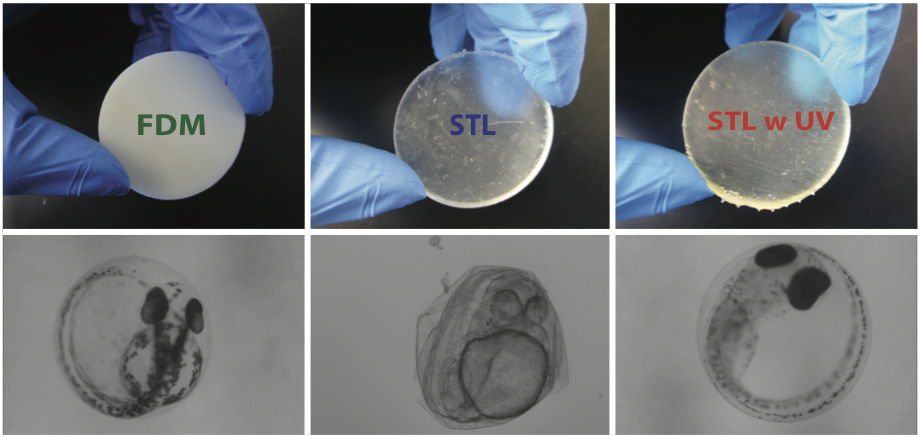
3D printing is gaining popularity by providing a tool for fast, cost-effective, and highly customizable fabrication. However, little is known about the toxicity of 3D-printed objects. In this work, we assess the toxicity of printed parts from two main classes of commercial 3D printers, fused deposition modeling and stereolithography. We assessed the toxicity of these 3D-printed parts using zebrafish (Danio rerio), a widely used model organism in aquatic toxicology. Zebrafish embryos were exposed to 3D-printed parts and monitored for rates of survival, hatching, and developmental abnormalities. We found that parts from both types of printers were measurably toxic to zebrafish embryos, with STL-printed parts significantly more toxic than FDM-printed parts. We also developed a simple post-printing treatment (exposure to ultraviolet light) that largely mitigates the toxicity of the STL-printed parts. Our results call attention to the need for strategies for the safe disposal of 3D-printed parts and printer waste materials.
-
Measuring single-cell density
William H. Grover, Andrea K. Bryan, Monica Diez-Silva, Subra Suresh, John M. Higgins, and Scott R. Manalis, Proceedings of the National Academy of Sciences of the United States of America 108 (27), 10992-10996 (2011). PDF
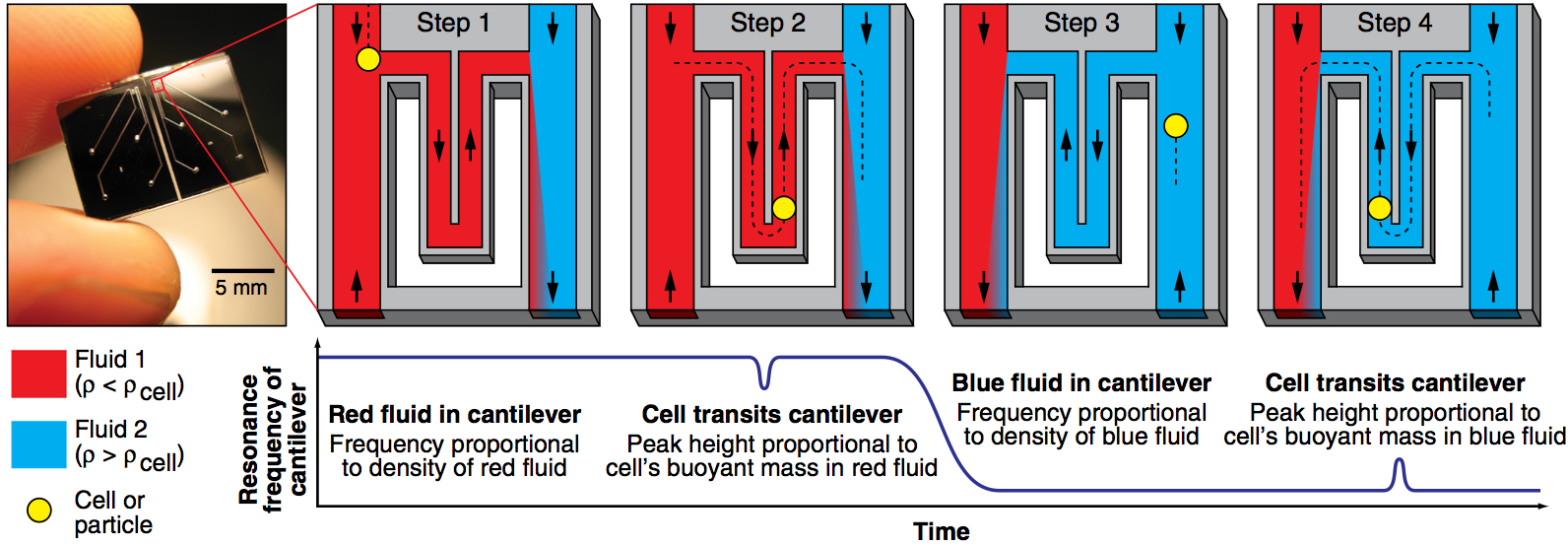
We have used a microfluidic mass sensor to measure the density of single living cells. By weighing each cell in two fluids of different densities, our technique measures the single-cell mass, volume, and density of approximately 500 cells per hour with a density precision of 0.001 g/mL. We observe that the intrinsic cell-to-cell variation in density is nearly 100-fold smaller than the mass or volume variation. As a result, we can measure changes in cell density indicative of cellular processes that would be otherwise undetectable by mass or volume measurements. Here we demonstrate this with four examples: identifying P. falciparum malaria-infected erythrocytes in a culture, distinguishing transfused blood cells from a patient’s own blood, identifying irreversibly-sickled cells in a sickle cell patient, and identifying leukemia cells in the early stages of responding to a drug treatment. These demonstrations suggest that the ability to measure single cell density will provide valuable insights into cell state for a wide range of biological processes.
-
Using buoyant mass to measure the growth of single cells
Michel Godin, Francisco Feijo Delgado, Sungmin Son, William H. Grover, Andrea K. Bryan, Amit Tzur, Paul Jorgensen, Kris Payer, Alan D. Grossman, Marc W. Kirschner, and Scott R. Manalis, Nature Methods 7, 387-390 (2010). PDF
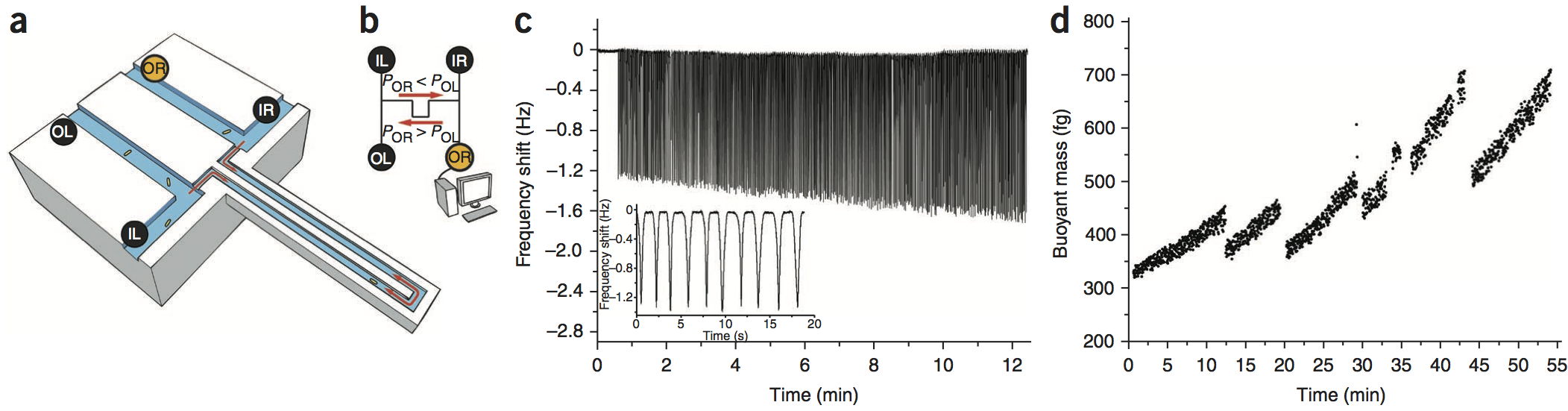
We used a suspended microchannel resonator (SMR) combined with picoliter-scale microfluidic control to measure buoyant mass and determine the “instantaneous” growth rates of individual cells. The SMR measures mass with femtogram precision, allowing rapid determination of the growth rate in a fraction of a complete cell cycle. We found that for individual cells of Bacillus subtilis, Escherichia coli, Saccharomyces cerevisiae and mouse lymphoblasts, heavier cells grew faster than lighter cells.
-
Teflon films for chemically-inert microfluidic valves and pumps
William H. Grover, Marcio G. von Muhlen, and Scott R. Manalis, Lab on a Chip 8 (6), 913-918 (2008). PDF
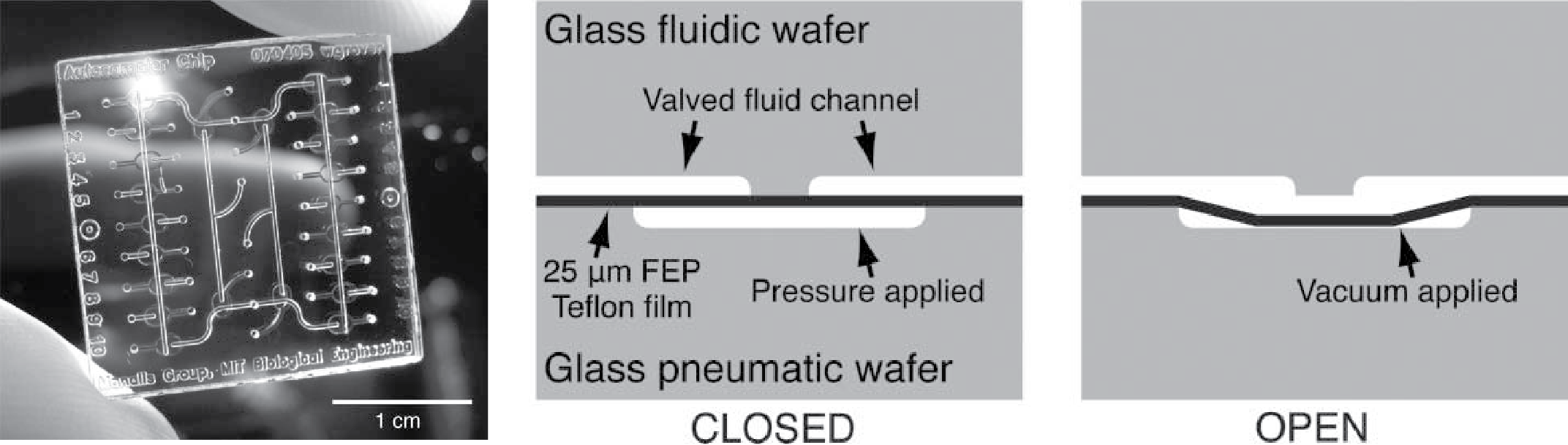
We present a simple method for fabricating chemically-inert Teflon microfluidic valves and pumps in glass microfluidic devices. These structures are modeled after monolithic membrane valves and pumps that utilize a featureless polydimethylsiloxane (PDMS) membrane bonded between two etched glass wafers. The limited chemical compatibility of PDMS has necessitated research into alternative materials for microfluidic devices. Previous work has shown that spin-coated amorphous fluoropolymers and Teflon-fluoropolymer laminates can be fabricated and substituted for PDMS in monolithic membrane valves and pumps for space flight applications. However, the complex process for fabricating these spin-coated Teflon films and laminates may preclude their use in many research and manufacturing contexts. As an alternative, we show that commercially-available fluorinated ethylene-propylene (FEP) Teflon films can be used to fabricate chemically-inert monolithic membrane valves and pumps in glass microfluidic devices. The FEP Teflon valves and pumps presented here are simple to fabricate, function similarly to their PDMS counterparts, maintain their performance over extended use, and are resistant to virtually all chemicals. These structures should facilitate lab-on-a-chip research involving a vast array of chemistries that are incompatible with native PDMS microfluidic devices.
-
Micropneumatic digital logic structures for integrated microdevice computation and control
Erik C. Jensen, William H. Grover, and Richard A. Mathies, Journal of Microelectromechanical Systems 16 (6), 1378-1385 (2007). PDF
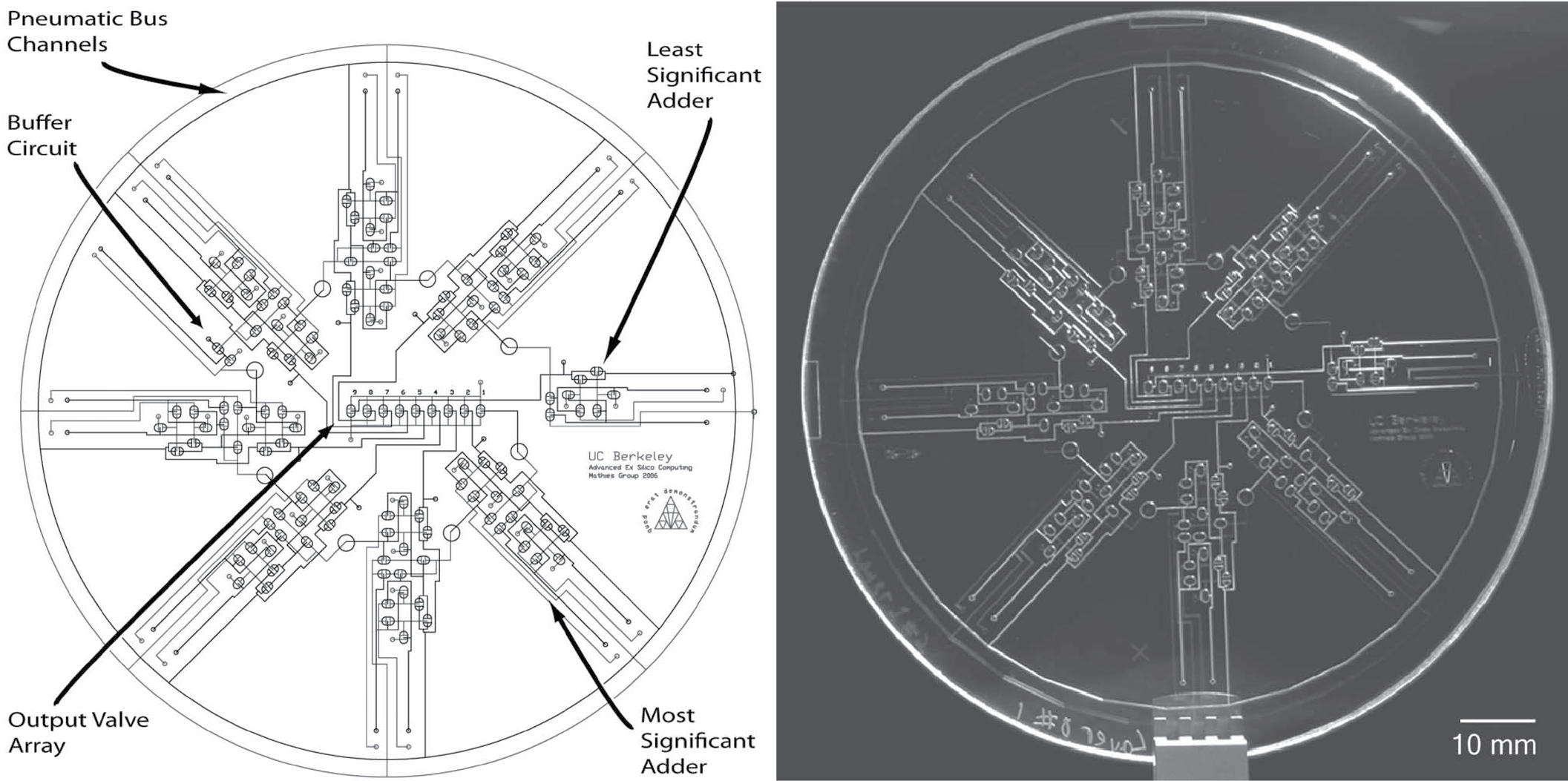
It is shown that microfabricated polydimethylsiloxane membrane valve structures can be configured to function as transistors in pneumatic digital logic circuits. Using the analogy with metal-oxide-semiconductor field-effect transistor circuits, networks of pneumatically actuated microvalves are designed to produce pneumatic digital logic gates (AND, OR, NOT, NAND, and XOR). These logic gates are combined to form 4- and 8-bit ripple-carry adders as a demonstration of their universal pneumatic computing capabilities. Signal propagation through these pneumatic circuits is characterized, and an amplifier circuit is demonstrated for improved signal transduction. Propagation of pneumatic carry information through the 8-bit adder is complete within 1.1 s, demonstrating the feasibility of integrated temporal control of pneumatic actuation systems. Integrated pneumatic logical systems reduce the number of off-chip controllers required for lab-on-a-chip and microelectromechanical system devices, allowing greater complexity and portability. This technology also enables the development of digital pneumatic computing and logic systems that are immune to electromagnetic interference.
-
Microfluidic serial dilution circuit
Brian M. Paegel, William H. Grover, Alison M. Skelley, Richard A. Mathies, Gerald F. Joyce, Analytical Chemistry 78 (21), 7522-7527 (2006). PDF
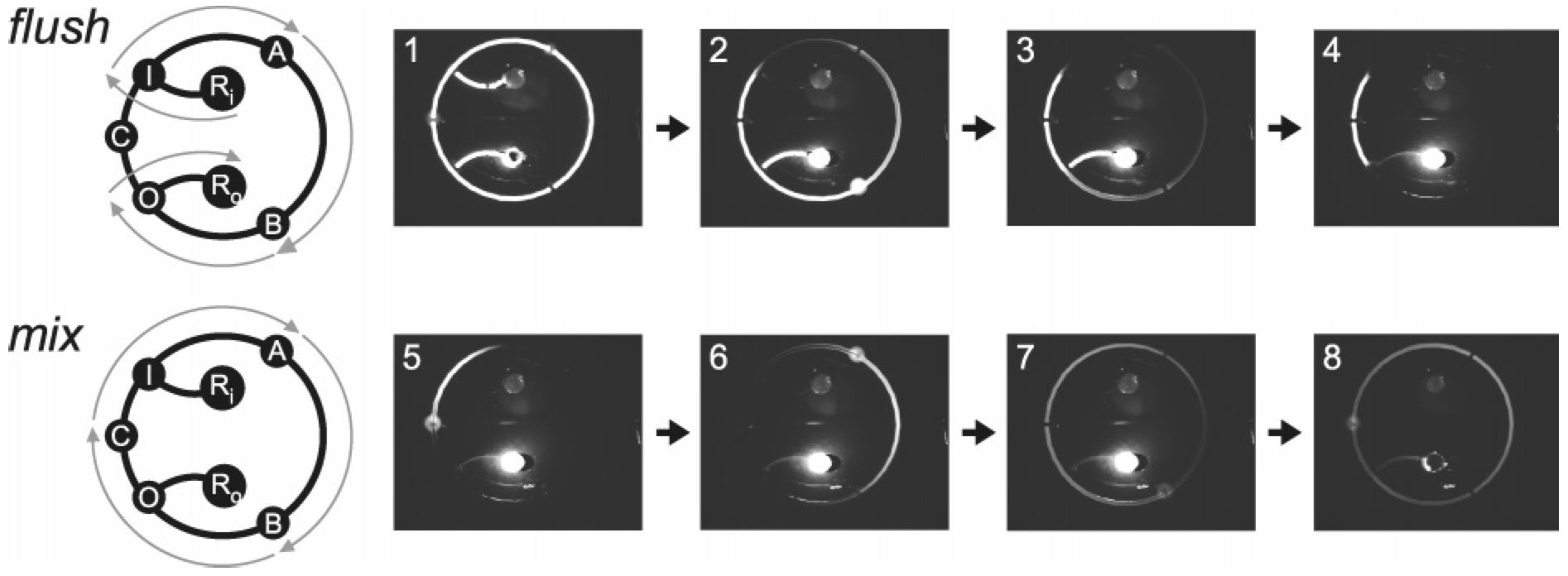
In vitro evolution of RNA molecules requires a method for executing many consecutive serial dilutions. To solve this problem, a microfluidic circuit has been fabricated in a three-layer glass-PDMS-glass device. The 400-nL serial dilution circuit contains five integrated membrane valves: three two-way valves arranged in a loop to drive cyclic mixing of the diluent and carryover, and two bus valves to control fluidic access to the circuit through input and output channels. By varying the valve placement in the circuit, carryover fractions from 0.04 to 0.2 were obtained. Each dilution process, which is composed of a diluent flush cycle followed by a mixing cycle, is carried out with no pipetting, and a sample volume of 400 nL is sufficient for conducting an arbitrary number of serial dilutions. Mixing is precisely controlled by changing the cyclic pumping rate, with a minimum mixing time of 22 s. This microfluidic circuit is generally applicable for integrating automated serial dilution and sample preparation in almost any microfluidic architecture.
-
Development and multiplexed control of latching pneumatic valves using microfluidic logical structures
William H. Grover, Robin H.C. Ivester, Erik C. Jensen, and Richard A. Mathies, Lab on a Chip 6 (5), 623-631 (2006). PDF
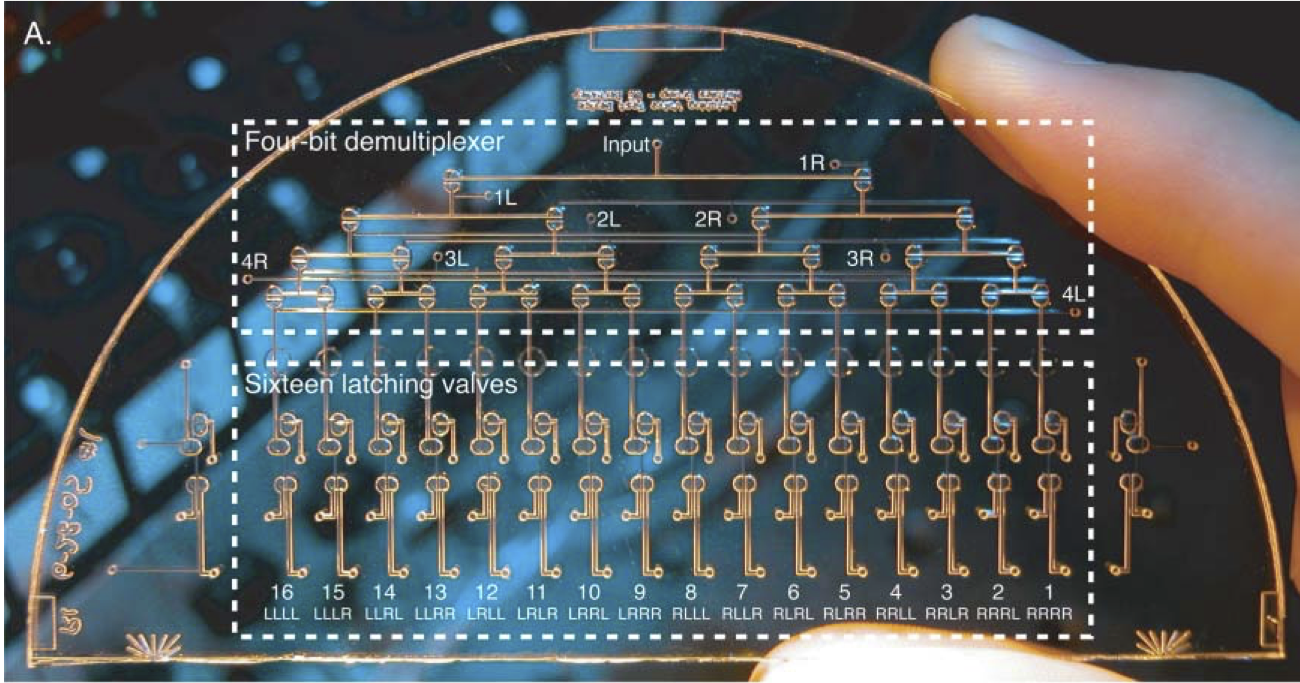
Novel latching microfluidic valve structures are developed, characterized, and controlled independently using an on-chip pneumatic demultiplexer. These structures are based on pneumatic monolithic membrane valves and depend upon their normally-closed nature. Latching valves consisting of both three- and four-valve circuits are demonstrated. Vacuum or pressure pulses as short as 120 ms are adequate to hold these latching valves open or closed for several minutes. In addition, an on-chip demultiplexer is demonstrated that requires only n pneumatic inputs to control 2(n-1) independent latching valves. These structures can reduce the size, power consumption, and cost of microfluidic analysis devices by decreasing the number of off-chip controllers. Since these valve assemblies can form the standard logic gates familiar in electronic circuit design, they should be useful in developing complex pneumatic circuits.
-
An integrated microfluidic processor for single nucleotide polymorphism-based DNA computing
William H. Grover and Richard A. Mathies, Lab on a Chip 5 (10), 1033-1040 (2005). PDF
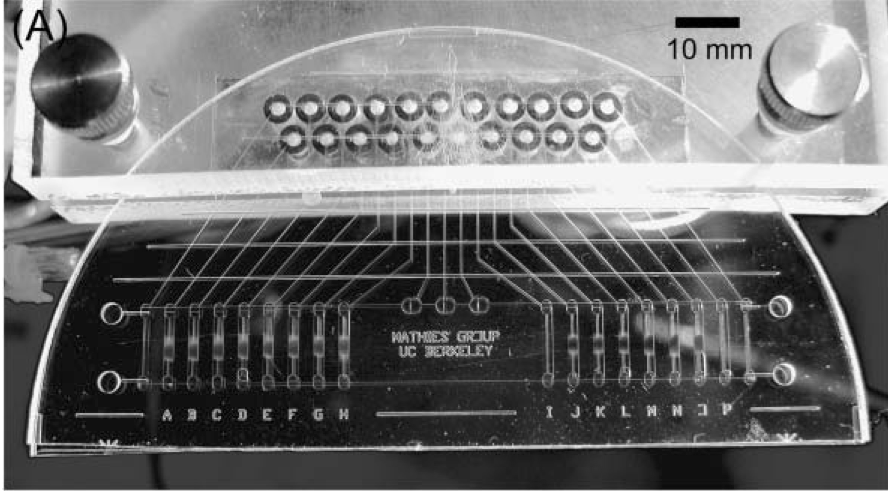
An integrated microfluidic processor is developed that performs molecular computations using single nucleotide polymorphisms (SNPs) as binary bits. A complete population of fluorescein-labeled DNA “answers” is synthesized containing three distinct polymorphic bases; the identity of each base (A or T) is used to encode the value of a binary bit (TRUE or FALSE). Computation and readout occur by hybridization to complementary capture DNA oligonucleotides bound to magnetic beads in the microfluidic device. Beads are loaded into sixteen capture chambers in the processor and suspended in place by an external magnetic field. Integrated microfluidic valves and pumps circulate the input DNA population through the bead suspensions. In this example, a program consisting of a series of capture/rinse/release steps is executed and the DNA molecules remaining at the end of the computation provide the solution to a three-variable, four-clause Boolean satisfiability problem. The improved capture kinetics, transfer efficiency, and single-base specificity enabled by microfluidics make our processor well-suited for performing larger-scale DNA computations.
-
Development and evaluation of a microdevice for amino acid biomarker detection and analysis on Mars
Alison M. Skelley, James R. Scherer, Andrew D. Aubrey, William H. Grover, Robin H.C. Ivester, Pascale Ehrenfreund, Frank J. Grunthaner, Jeffrey L. Bada, Richard A. Mathies, Proceedings of the National Academy of Sciences of the United States of America 102 (4), 1041-1046 (2005). PDF
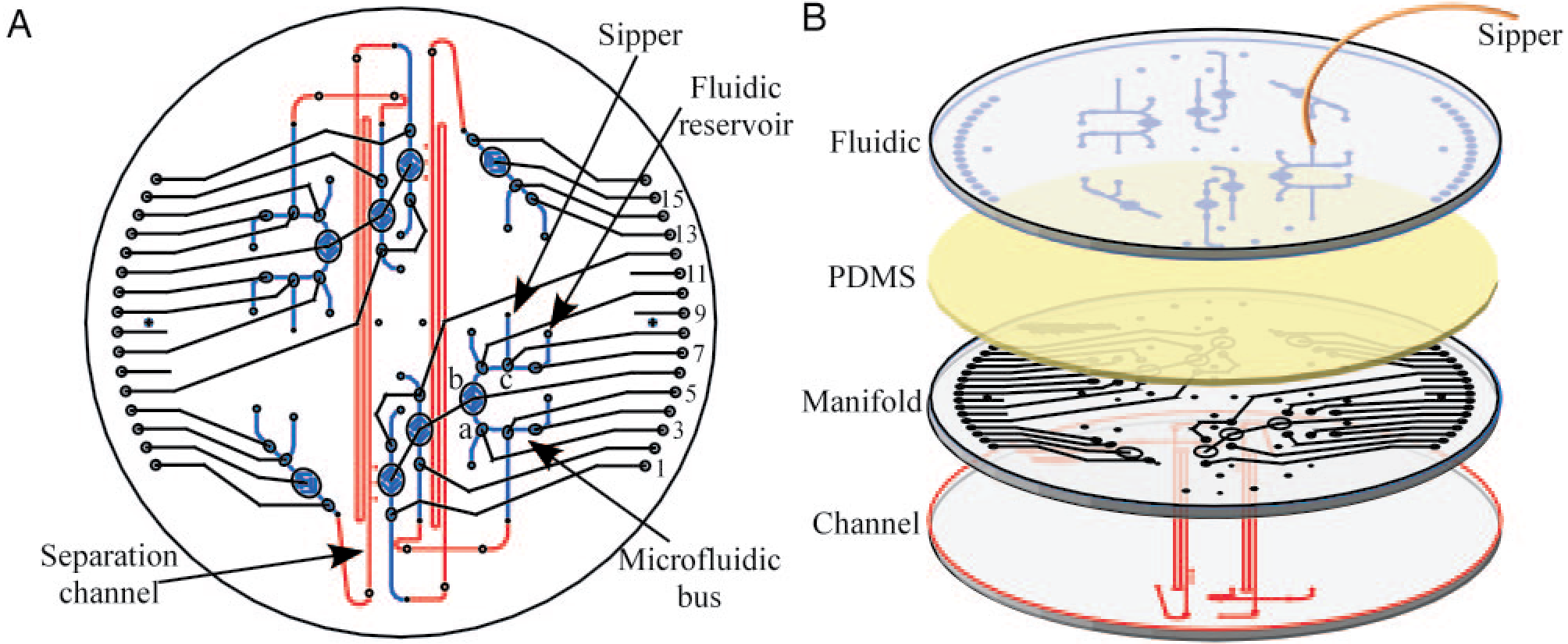
The Mars Organic Analyzer (MOA), a microfabricated capillary electrophoresis (CE) instrument for sensitive amino acid biomarker analysis, has been developed and evaluated. The microdevice consists of a four-wafer sandwich combining glass CE separation channels, microfabricated pneumatic membrane valves and pumps, and a nanoliter fluidic network. The portable MOA instrument integrates high voltage CE power supplies, pneumatic controls, and fluorescence detection optics necessary for field operation. The amino acid concentration sensitivities range from micromolar to 0.1 nM, corresponding to part-per-trillion sensitivity. The MOA was first used in the lab to analyze soil extracts from the Atacama Desert, Chile, detecting amino acids ranging from 10-600 parts per billion. Field tests of the MOA in the Panoche Valley, CA, successfully detected amino acids at 70 parts per trillion to 100 parts per billion in jarosite, a sulfate-rich mineral associated with liquid water that was recently detected on Mars. These results demonstrate the feasibility of using the MOA to perform sensitive in situ amino acid biomarker analysis on soil samples representative of a Mars-like environment.
-
Monolithic membrane valves and diaphragm pumps for practical large-scale integration into glass microfluidic devices
William H. Grover, Alison M. Skelley, Chung N. Liu, Eric T. Lagally, Richard A. Mathies, Sensors and Actuators B 89 (3), 315-323 (2003). PDF
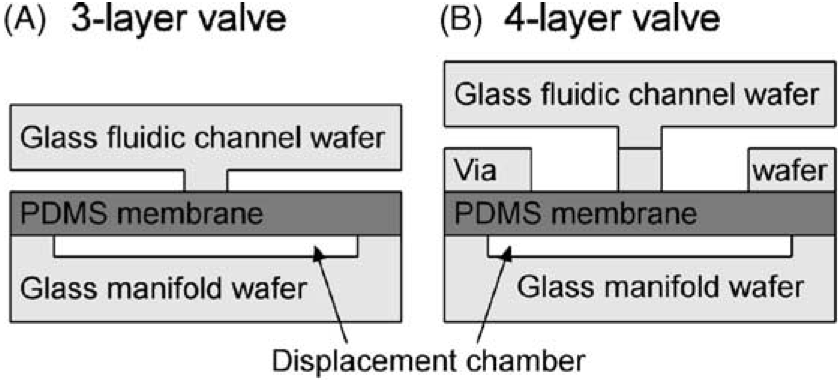
Monolithic elastomer membrane valves and diaphragm pumps suitable for large-scale integration into glass microfluidic analysis devices are fabricated and characterized. Valves and pumps are fabricated by sandwiching an elastomer membrane between etched glass fluidic channel and manifold wafers. A three-layer valve and pump design features simple non-thermal device bonding and a hybrid glass-PDMS fluidic channel; a four-layer structure includes a glass fluidic system with minimal fluid-elastomer contact for improved chemical and biochemical compatibility. The pneumatically actuated valves have less than 10 nl dead volumes, can be fabricated in dense arrays, and can be addressed in parallel via an integrated manifold. The membrane valves provide flow rates up to 380 nL/s at 30 kPa driving pressure and seal reliably against fluid pressures as high as 75 kPa. The diaphragm pumps are self-priming, pump from a few nanoliters to a few microliters per cycle at overall rates from 1 to over 100 nl/s, and can reliably pump against 42 kPa pressure heads. These valves and pumps provide a facile and reliable integrated technology for fluid manipulation in complex glass microfluidic and electrophoretic analysis devices.